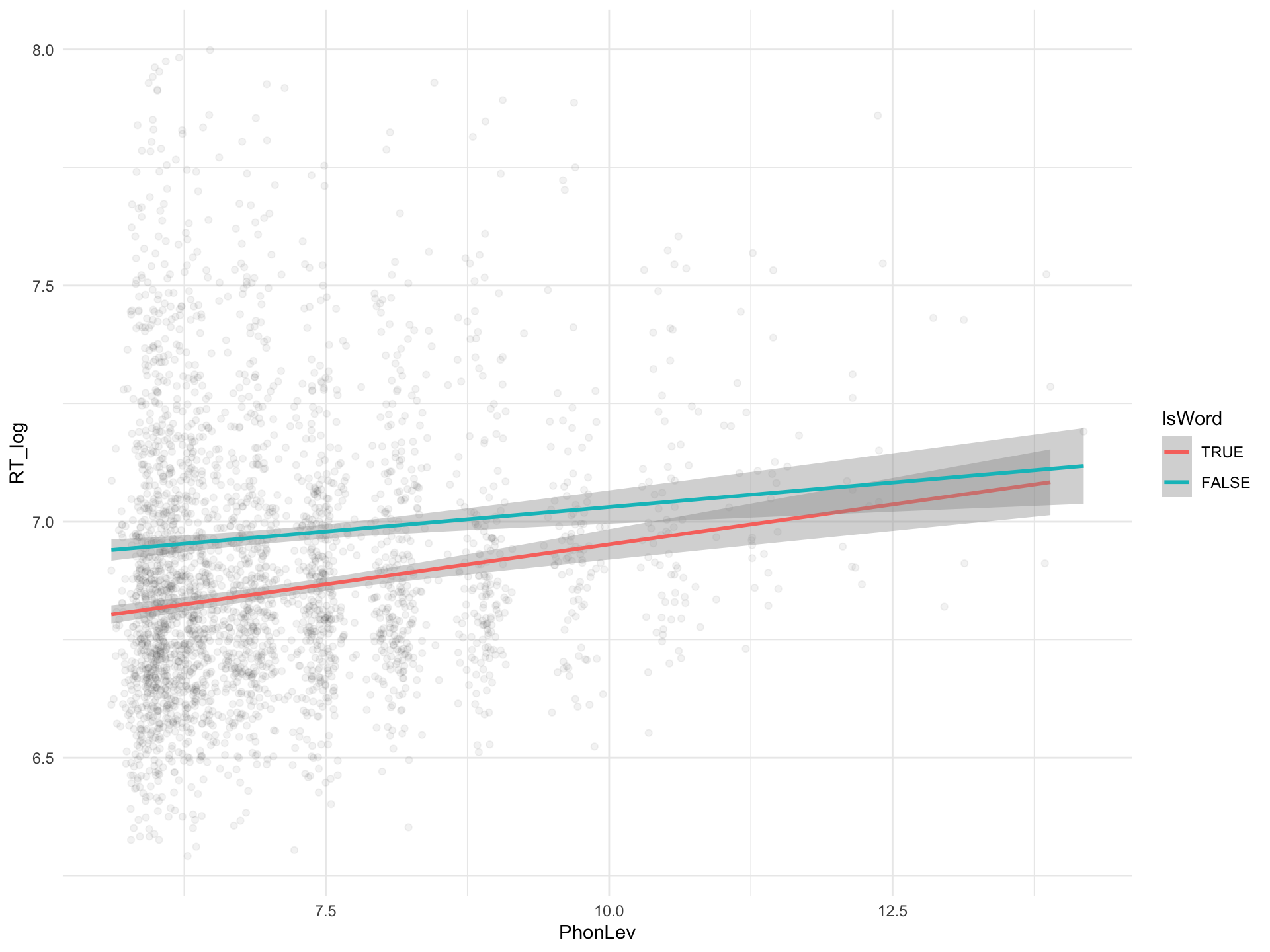
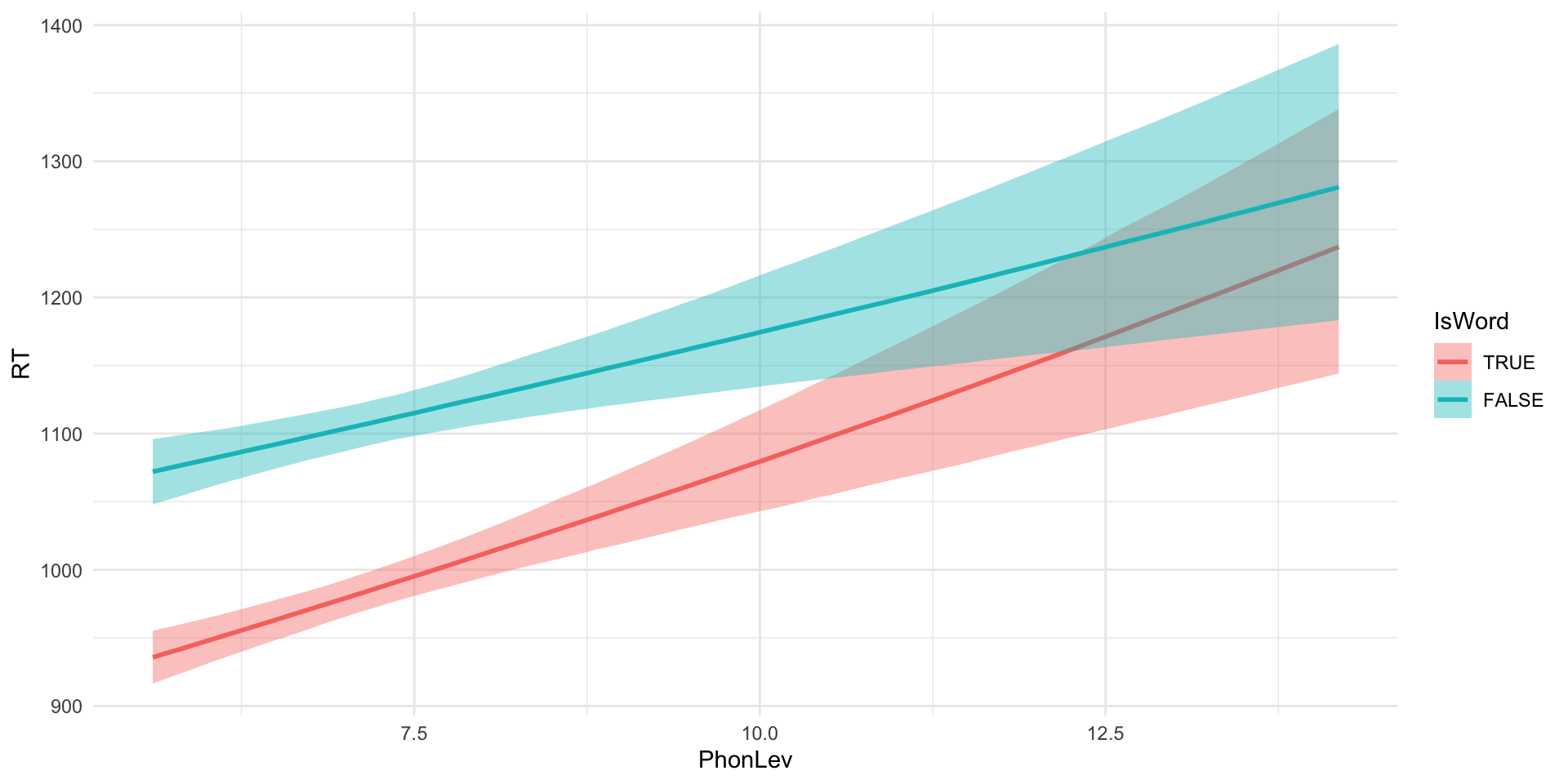
brm_int <- brm(
RT ~ 0 + IsWord + IsWord:PhonLev,
data = mald,
family = lognormal,
cores = 4,
seed = 9812,
file = "data/cache/brm_int.rds"
)
brm_int Family: lognormal
Links: mu = identity; sigma = identity
Formula: RT ~ 0 + IsWord + IsWord:PhonLev
Data: mald (Number of observations: 3000)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
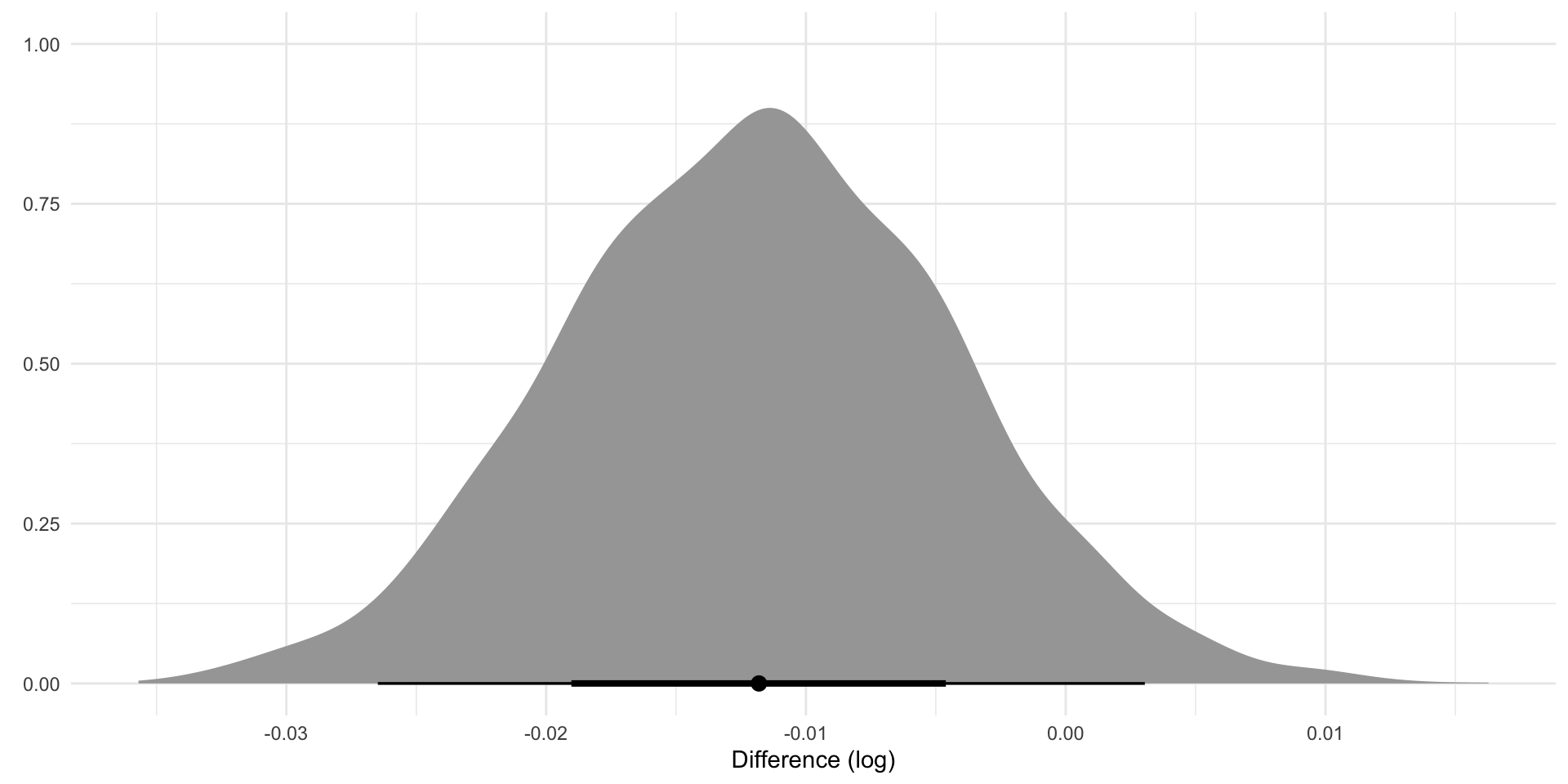
IsWordTRUE 6.62 0.04 6.54 6.70 1.00 1888 1924
IsWordFALSE 6.82 0.04 6.74 6.90 1.00 1414 1810
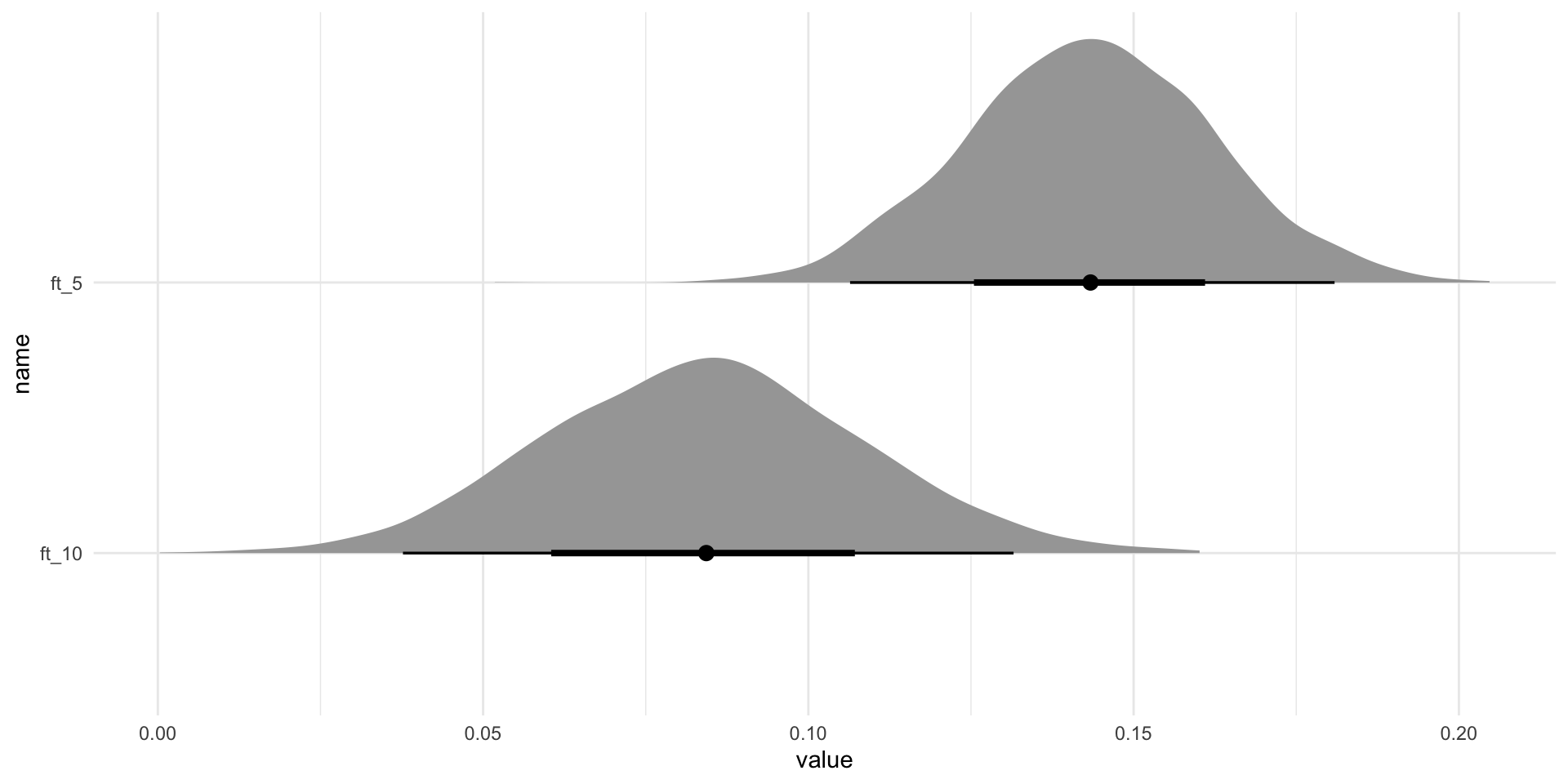
IsWordTRUE:PhonLev 0.03 0.01 0.02 0.04 1.00 1890 1877
IsWordFALSE:PhonLev 0.02 0.01 0.01 0.03 1.00 1438 1791
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.28 0.00 0.27 0.29 1.00 2673 2072
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).