Introduction to Bayesian regression models
03 - Diagnostics
University of Edinburgh
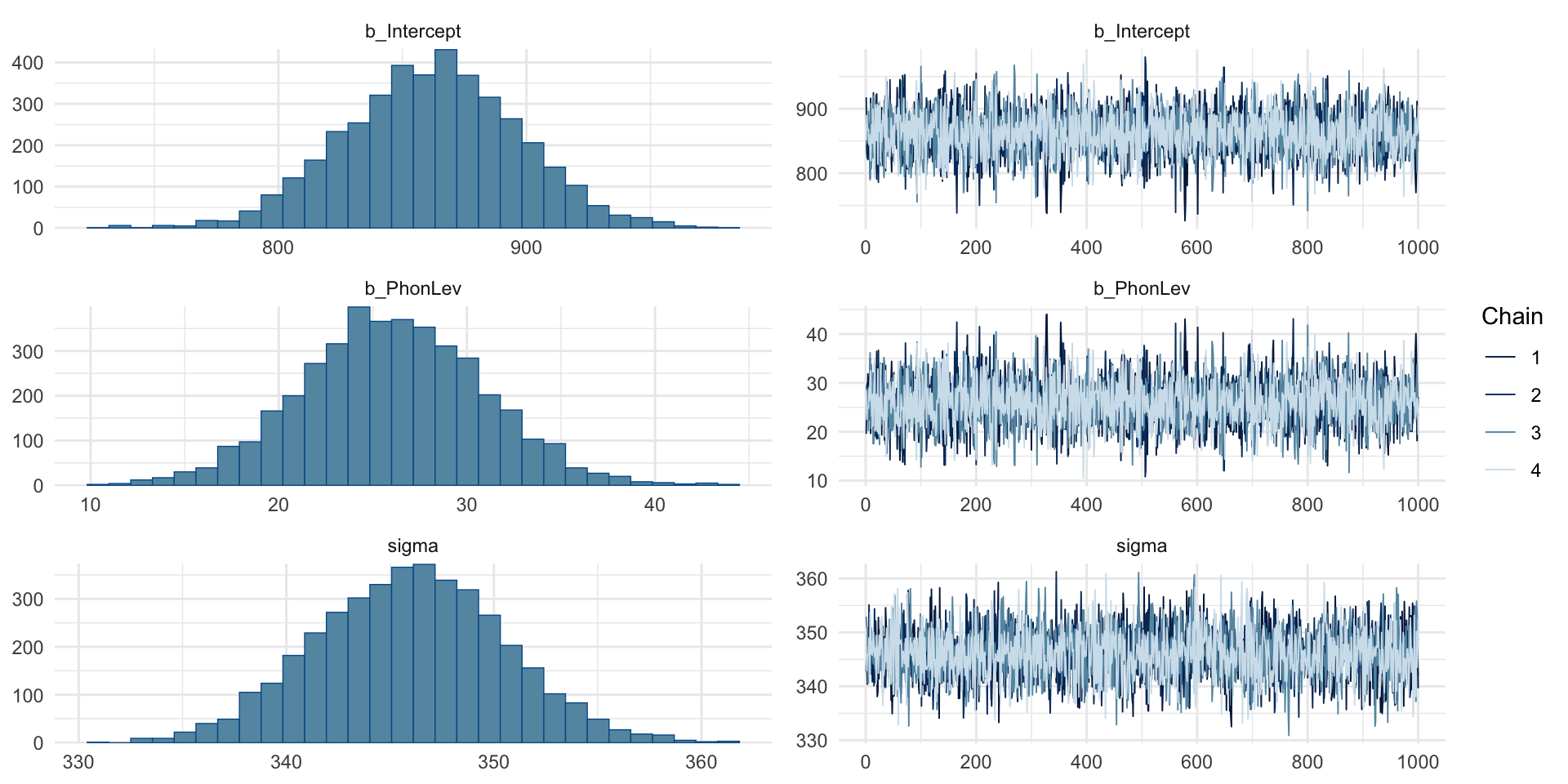
MCMC traces
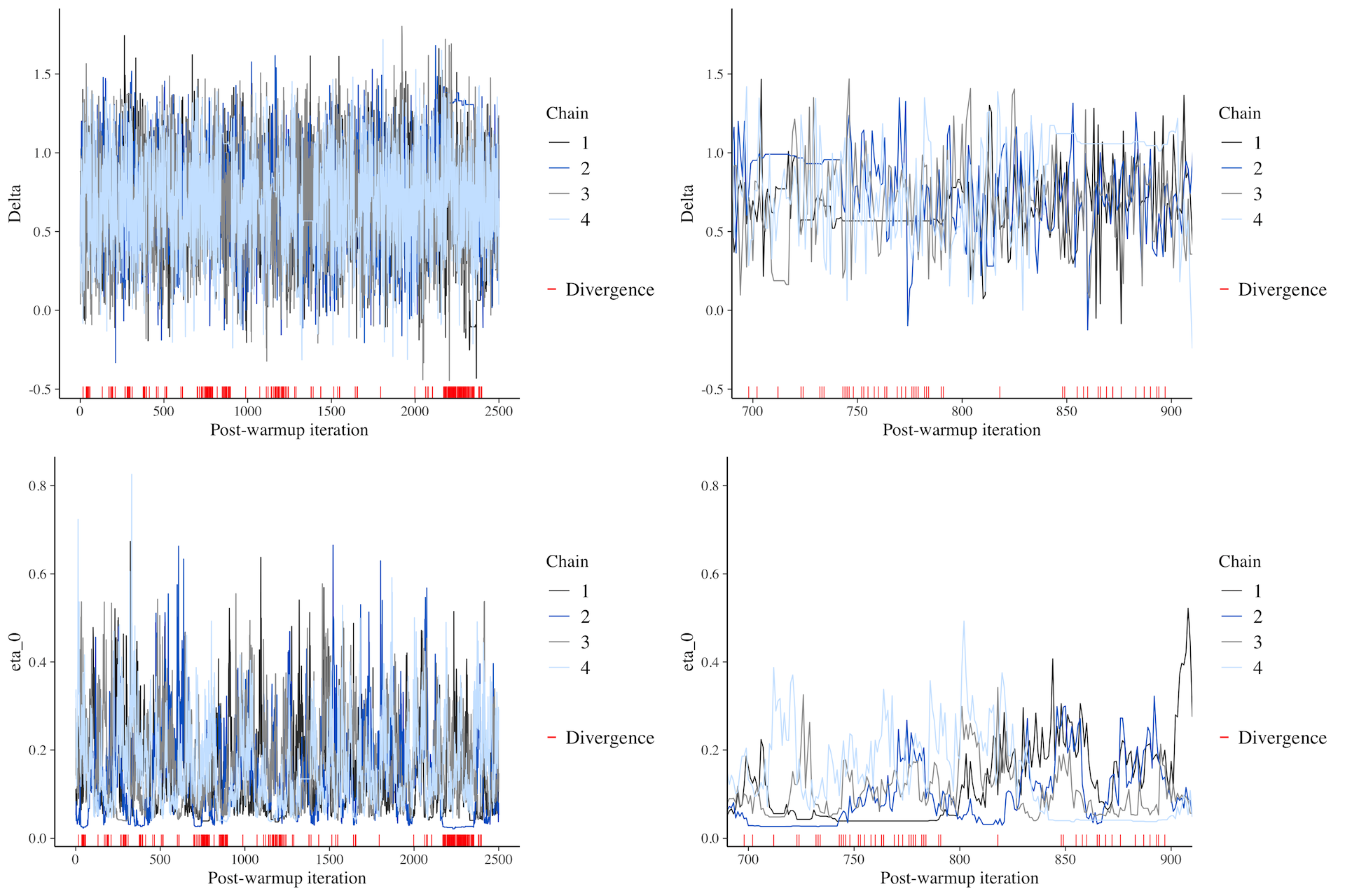
MCMC traces: bad
An example of bad MCMC chain mixing.
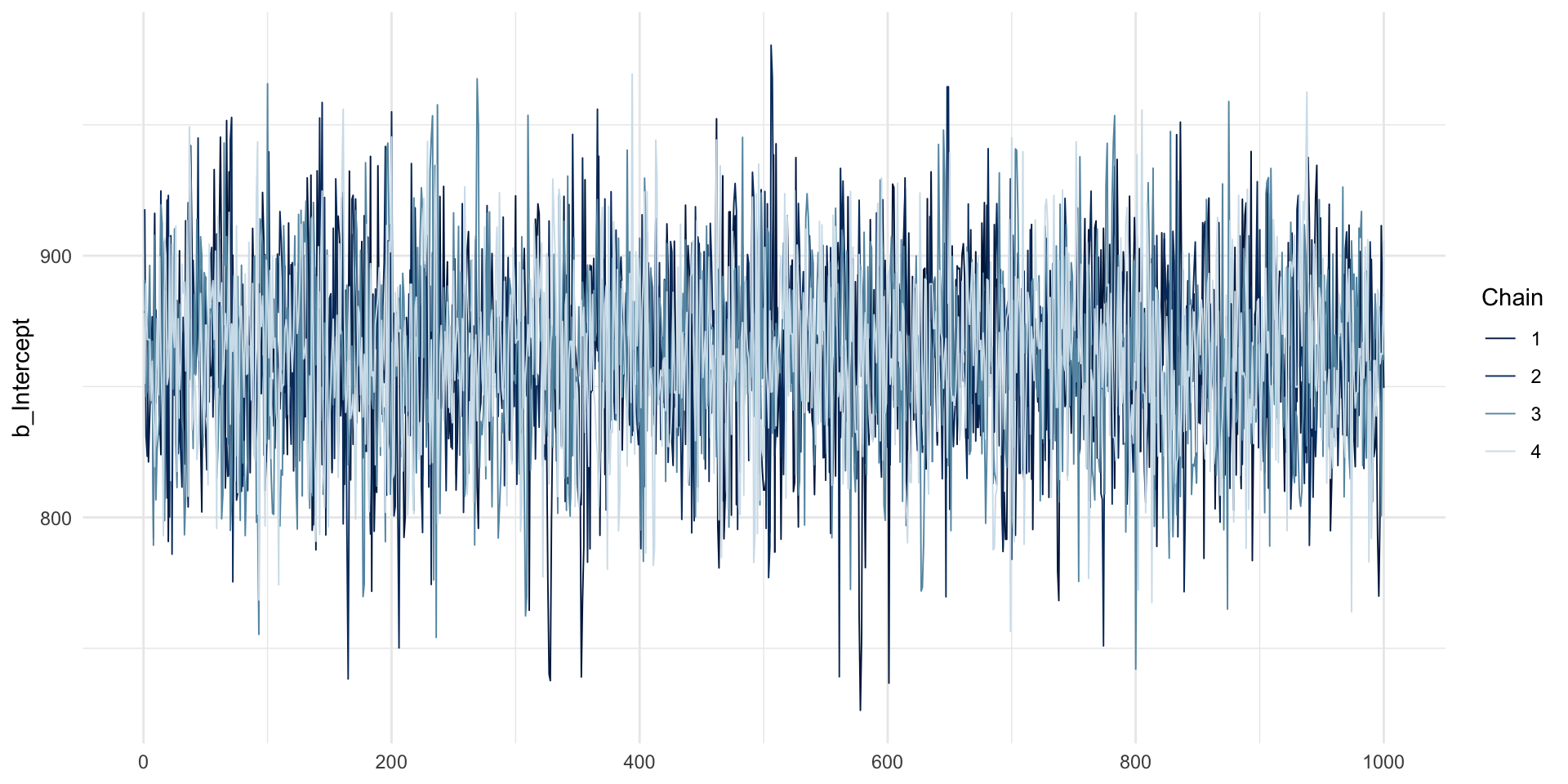
MCMC traces: intercept
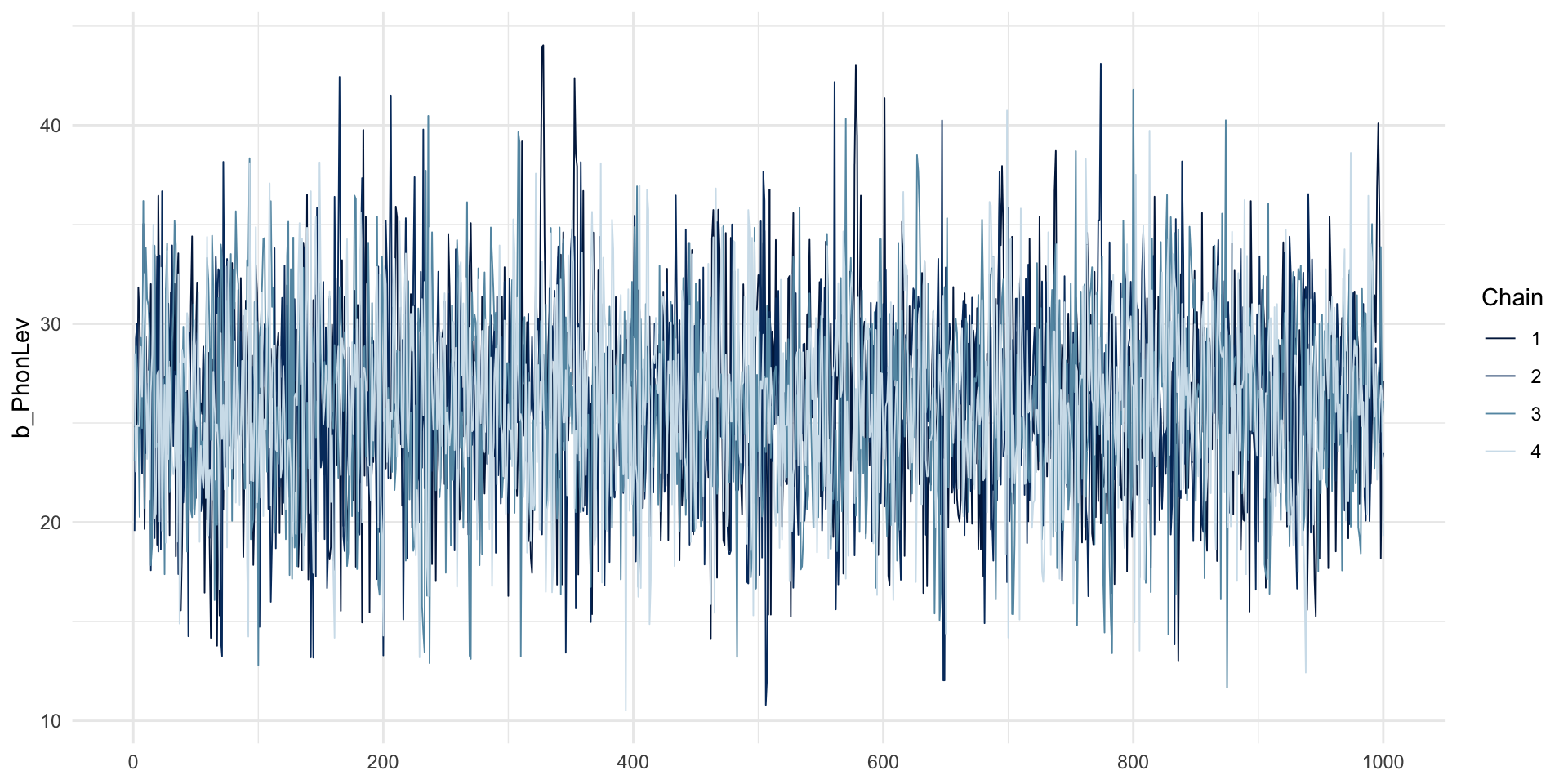
MCMC traces: IsWord
\(\hat{R}\) and Effective Sample Size (ESS)
Family: gaussian
Links: mu = identity; sigma = identity
Formula: RT ~ 1 + PhonLev
Data: mald (Number of observations: 3000)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 861.62 35.11 793.25 929.05 1.00 4139 2815
PhonLev 26.05 4.85 16.70 35.40 1.00 4169 2866
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 345.95 4.54 337.09 354.71 1.00 3849 2919
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).brms warns you
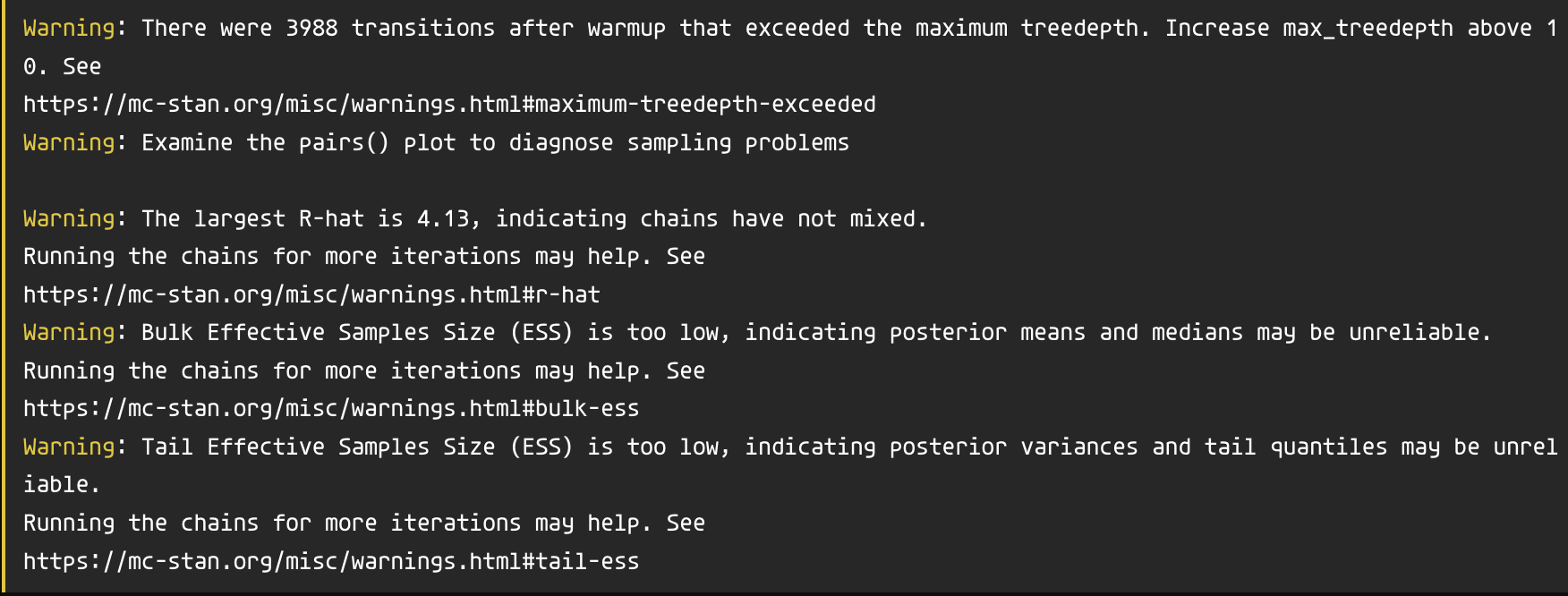
What to do
Increase the number of iterations (default is 2000):
iter = 4000.Increase
adapt_delta(an MCMC setting, default is 0.9, can only be between 0 and 1 exclusive).control = list(adapt_delta = 0.9999)
Increase
max_treedepth(another MCMC settings, default is 10).control = list(max_treedepth = 15)
These solutions increase the time needed to fit the model, which is a perfectly acceptable compromise.
Fictitious example
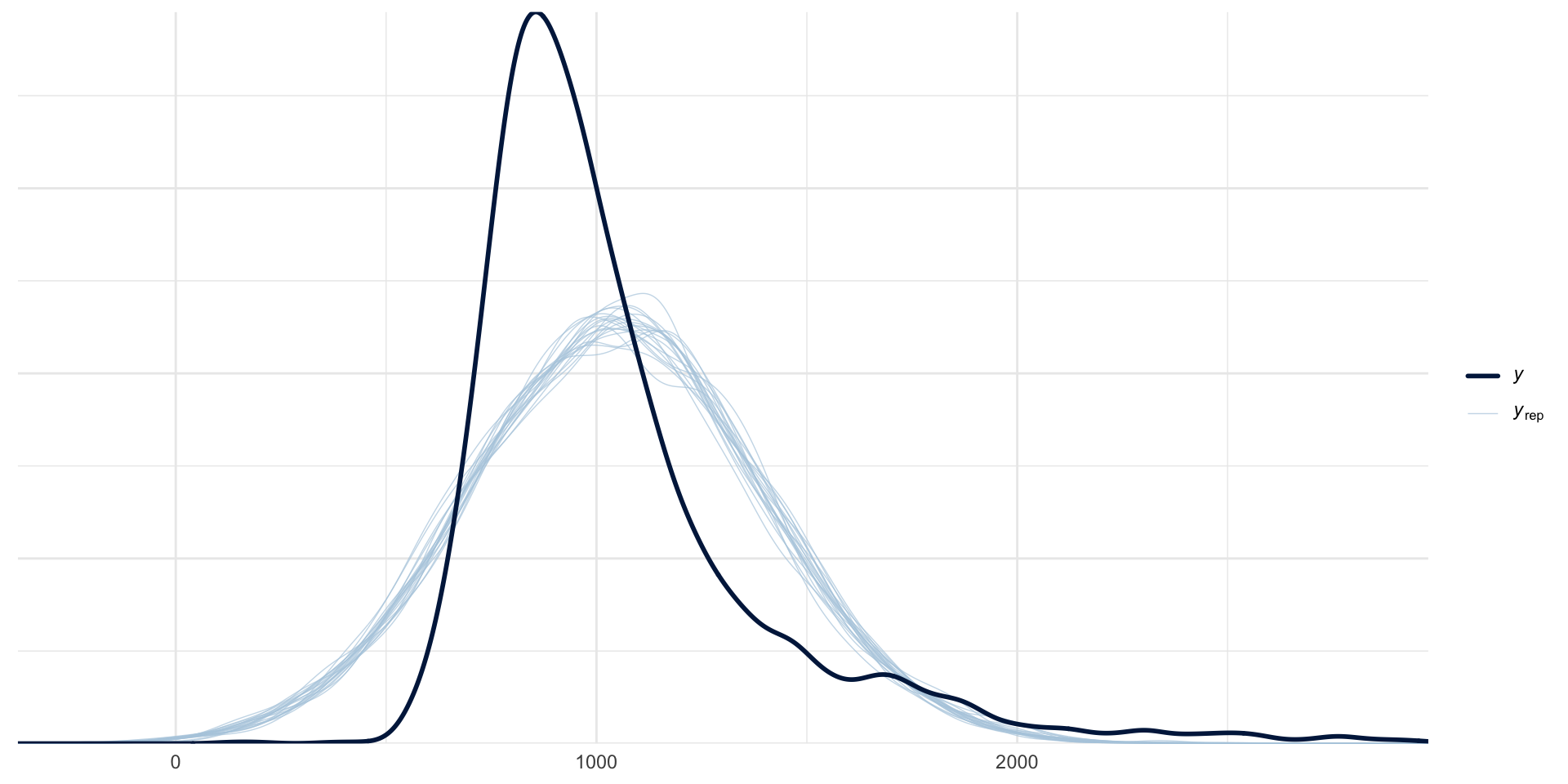
Posterior Predictive Checks
Log-normal regression
\[ \begin{align} RT & \sim Lognormal(\mu, \sigma)\\ log(\mu) & = \beta_0 + \beta_1 \cdot \text{PhonLev}\\ \end{align} \]
Variables that can only be positive, like Reaction Times, cannot be Gaussian.
A standard distribution for these variables is the log-normal distribution.
Log-normal regression: MALD
Log-normal regression: summary
Family: lognormal
Links: mu = identity; sigma = identity
Formula: RT ~ 1 + PhonLev
Data: mald (Number of observations: 3000)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 6.71 0.03 6.66 6.76 1.00 4948 3345
PhonLev 0.03 0.00 0.02 0.04 1.00 4954 3380
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.28 0.00 0.28 0.29 1.00 2395 2781
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
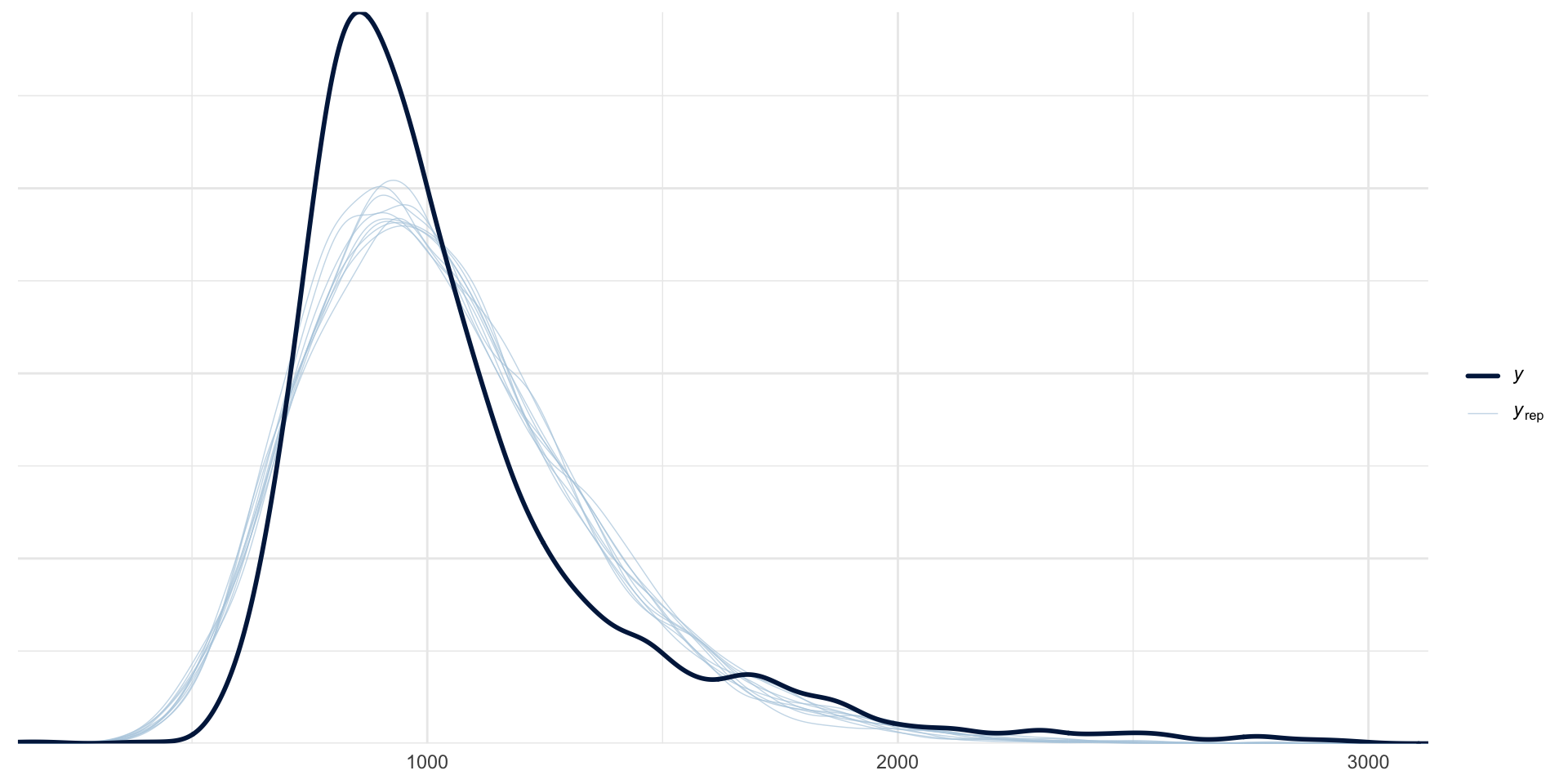
scale reduction factor on split chains (at convergence, Rhat = 1).Posterior Predictive Checks: looks better!
Predicted RTs by PhonLev
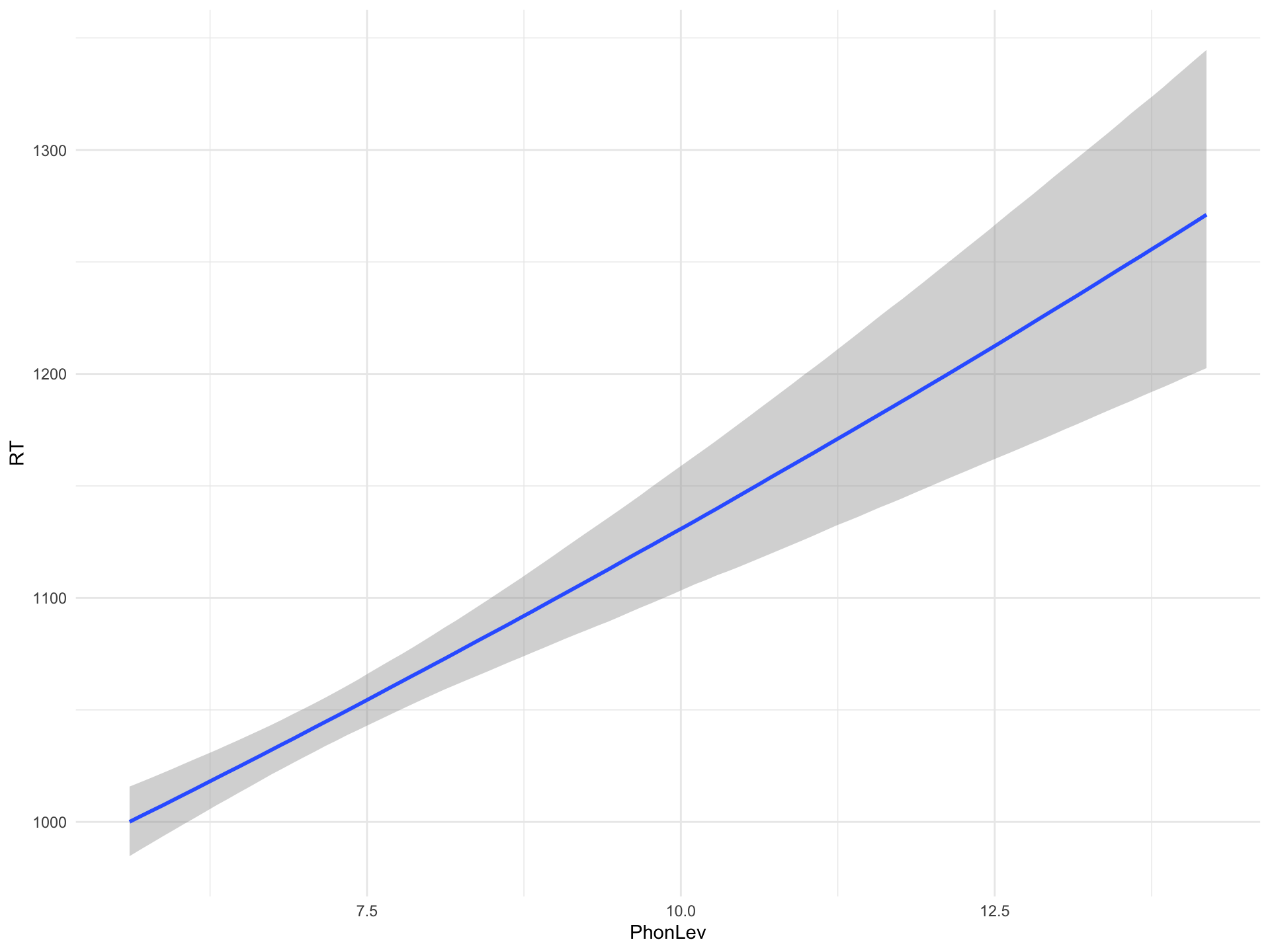
Figure 1: Predicted effect of mean phone-level distance on RTs from a Bayesian regression model.
Predicted RTs at representative values of PhonLev
Calculating CrIs from the draws
library(posterior)
brm_log_cri <- brm_log_draws |>
select(rt_05, rt_10, rt_15) |>
pivot_longer(everything()) |>
group_by(name) |>
summarise(
# Use quantile2() from posterior package
lo_95 = round(quantile2(value, 0.025)),
hi_95 = round(quantile2(value, 0.0975))
)
brm_log_cri# A tibble: 3 × 3
name lo_95 hi_95
<chr> <dbl> <dbl>
1 rt_05 926 932
2 rt_10 1059 1068
3 rt_15 1174 1200Summary
Quick and dirty diagnostics:
MCMC traces: hairy caterpillars, no divergent transitions.
\(\hat{R}\): should be 1 (> 1 means non-convergence).
Effective Sample Size (ESS): should be large enough.
Posterior Predictive Checks: predicted outcome distribution should match the empirical distribution.
brm()warns you about divergent transition, \(\hat{R} > 1\) and low ESS and how to fix them.- This usually involves increasing the number of iterations and/or other MCMC tricks.
Posterior predictive checks are based on visual inspection only.