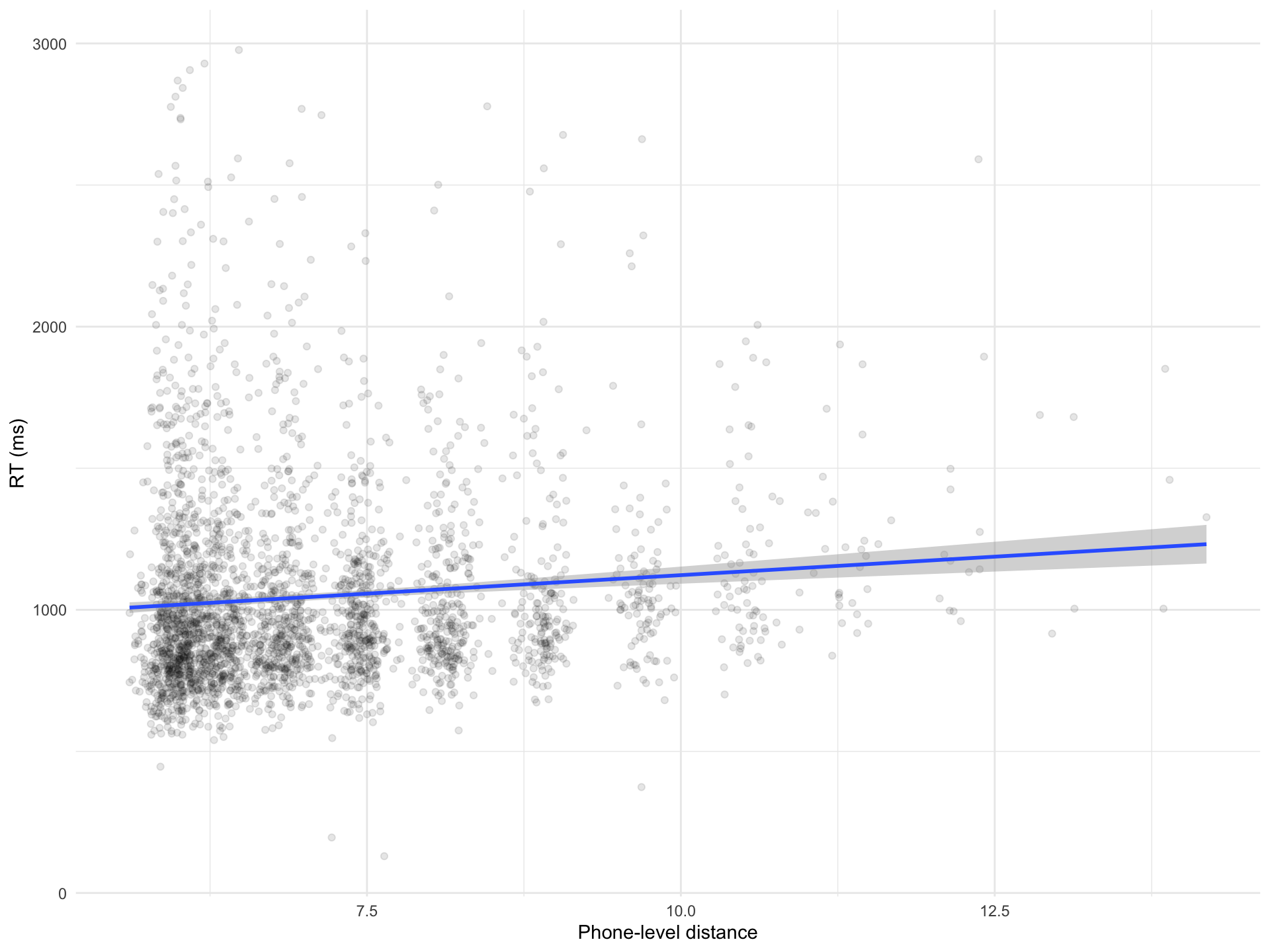
brm_lev <- brm(
RT ~ 1 + PhonLev,
data = mald,
family = "gaussian",
file = "./data/cache/brm_lev.rds",
chains = 4,
iter = 2000,
cores = 4
)
summary(brm_lev) Family: gaussian
Links: mu = identity; sigma = identity
Formula: RT ~ 1 + PhonLev
Data: mald (Number of observations: 3000)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
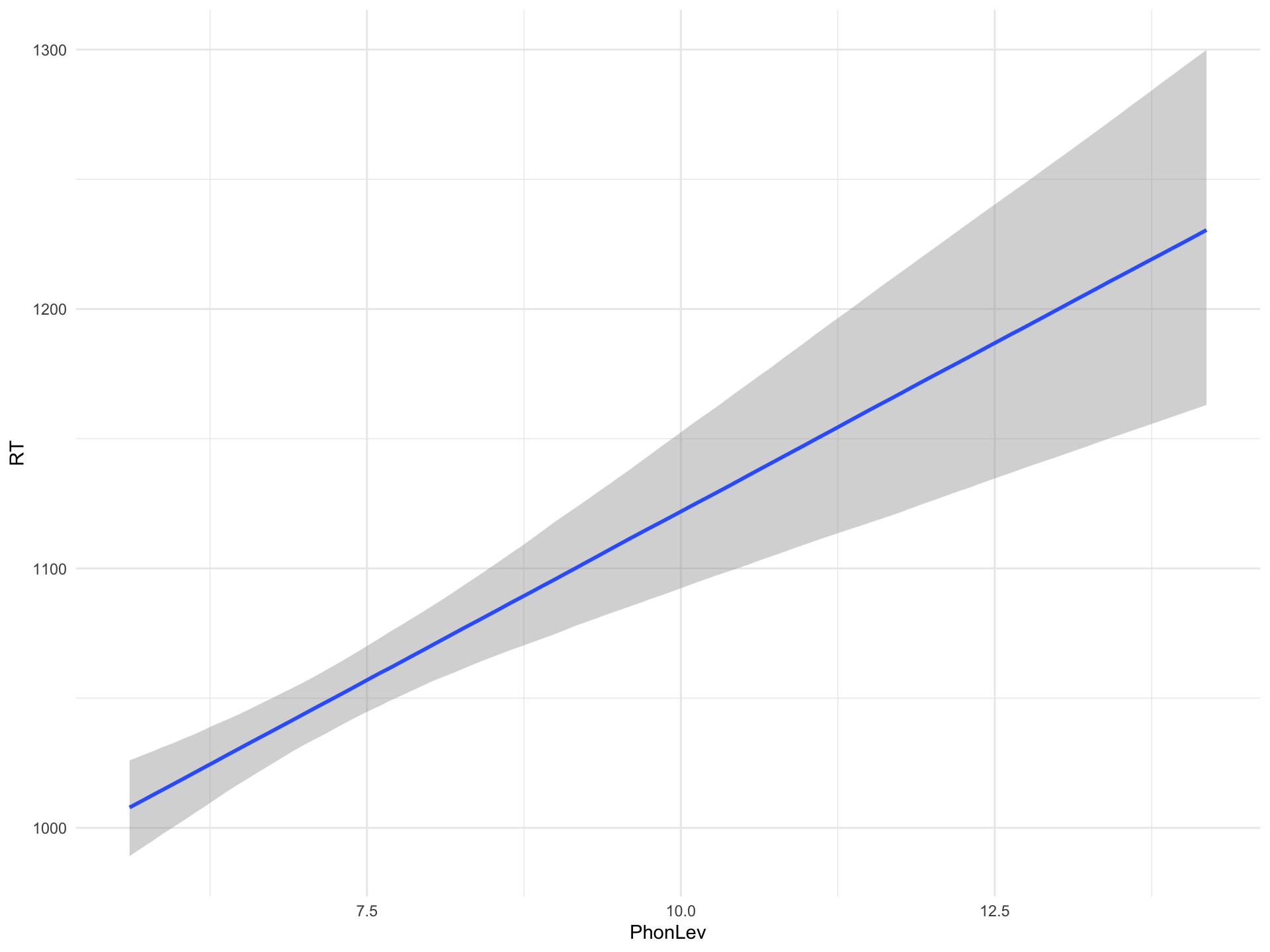
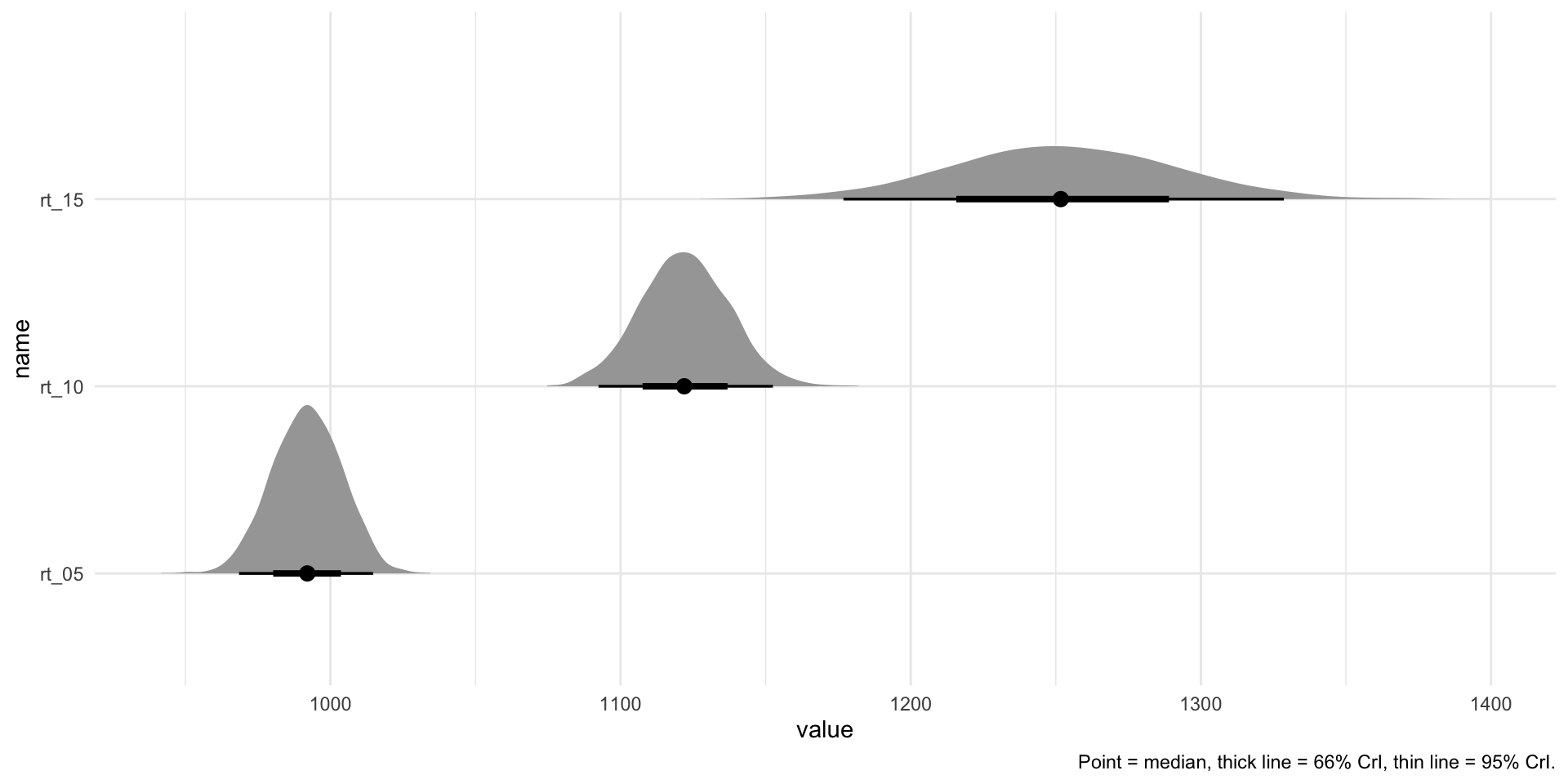
Intercept 861.62 35.11 793.25 929.05 1.00 4139 2815
PhonLev 26.05 4.85 16.70 35.40 1.00 4169 2866
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 345.95 4.54 337.09 354.71 1.00 3849 2919
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).