# A tibble: 3,000 × 7
Subject Item IsWord PhonLev RT ACC RT_log
<chr> <chr> <fct> <dbl> <int> <fct> <dbl>
1 15345 nihnaxr FALSE 5.84 945 correct 6.85
2 15345 skaep FALSE 6.03 1046 incorrect 6.95
3 15345 grandparents TRUE 10.3 797 correct 6.68
4 15345 sehs FALSE 5.88 2134 correct 7.67
5 15345 cousin TRUE 5.78 597 correct 6.39
6 15345 blowup TRUE 6.03 716 correct 6.57
7 15345 hhehrnzmaxn FALSE 7.30 1985 correct 7.59
8 15345 mantic TRUE 6.21 1591 correct 7.37
9 15345 notable TRUE 6.82 620 correct 6.43
10 15345 prowthihviht FALSE 7.68 1205 correct 7.09
# ℹ 2,990 more rowsIntroduction to Bayesian regression models
01 - Basics
University of Edinburgh
Download the workshop materials
- Download the workshop repository from: https://github.com/stefanocoretta/learnBayes.
Schedule
| Times | Topic |
|---|---|
| 9.00-10.30 | Introduction to Bayesian modelling |
| 10.30-11.00 | Break |
| 11.00-12.30 | Bayesian regression + Diagnostics |
| 12.30-14.30 | Lunch break |
| 14.30-16.00 | Categorical predictors + Interactions |
| 16.00-16.30 | Break |
| 16.30-18.00 | Bayesian modelling Q&A |

One model to rule them all.
One model to find them.
One model to bring them all
And in the darkness bind them.
(No, I couldn’t translate ‘model’ into Black Speech, alas…)
Now enter The Regression Model
Now enter The Bayesian Regression Model
All about the Bayes
Within the NHST (Frequentist) framework, the main analysis output is:
- Point estimates of predictors’ parameters (with standard error).
- p-values.
Within the Bayesian framework, the main analysis output is:
- Probability distributions of predictors’ parameters.
An example: MALD
Massive Auditory Lexical Decision (MALD) data (Tucker et al. 2019):
- Auditory Lexical Decision task with real and nonce English words.
- Reaction times and accuracy.
- Total 521 subjects; subset of 30 subjects, 100 observations each.
MALD data
MALD: phone-level distance and lexical status
Let’s investigate the effects of the following on Reaction Times (RTs):
PhonLev: mean phone-level Levenshtein distance of the item from all other entries.- An alternative to neighbourhood density.
IsWord: lexical status (real word vs nonce word).
MALD: phone-level distance and lexical status
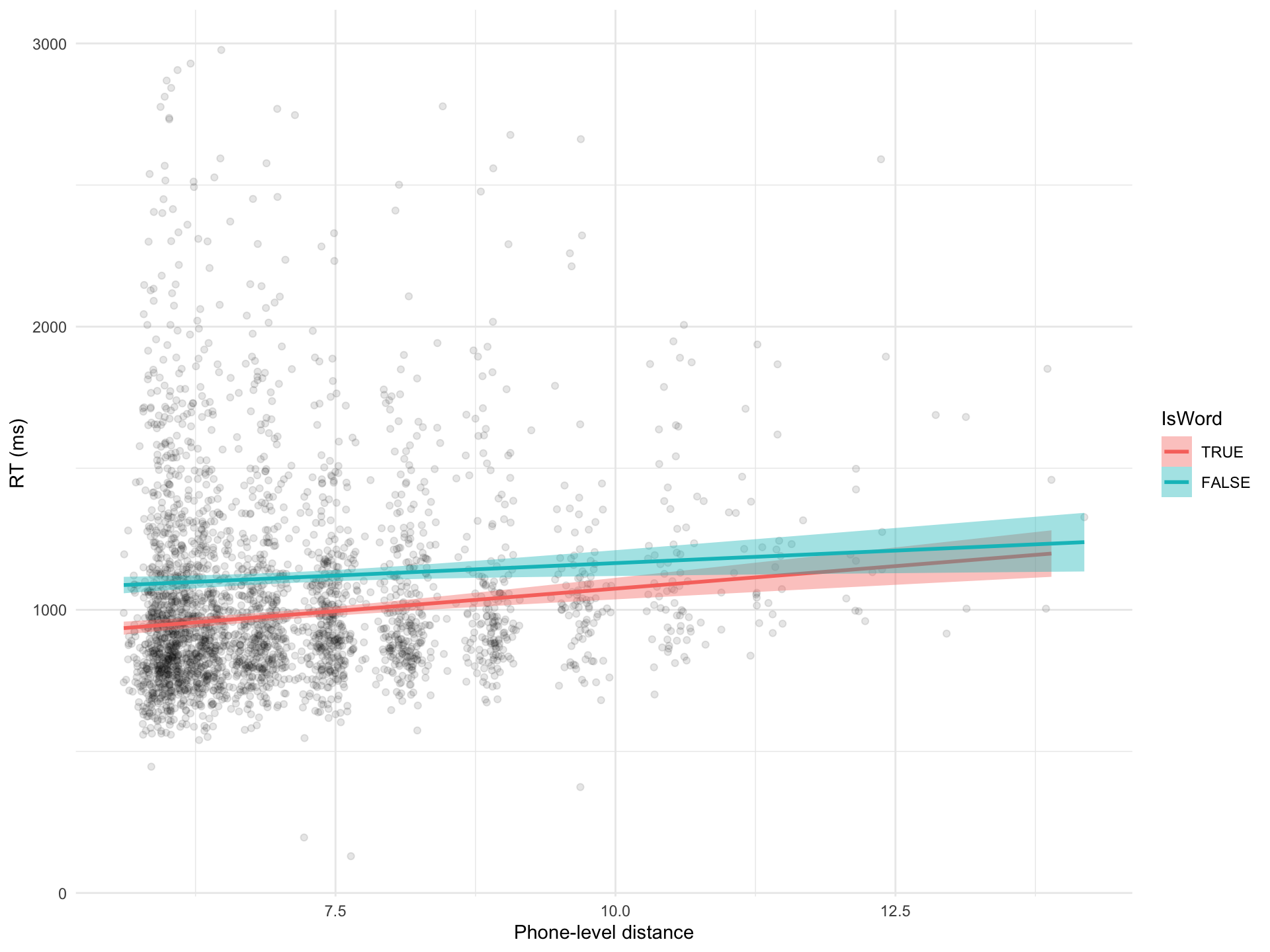
Figure 1: Scatter plot of mean phone-level distance vs reaction times.
A frequentist regression model
A frequentist regression model: summary
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: RT ~ PhonLev + IsWord + PhonLev:IsWord + (1 | Subject)
Data: mald
REML criterion at convergence: 43232.4
Scaled residuals:
Min 1Q Median 3Q Max
-3.5511 -0.5985 -0.2439 0.3185 5.6906
Random effects:
Groups Name Variance Std.Dev.
Subject (Intercept) 11032 105.0
Residual 104648 323.5
Number of obs: 3000, groups: Subject, 30
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 754.965 49.687 862.751 15.195 < 2e-16 ***
PhonLev 32.148 6.389 2970.933 5.032 5.15e-07 ***
IsWordFALSE 212.440 65.453 2972.588 3.246 0.00118 **
PhonLev:IsWordFALSE -11.620 9.076 2972.817 -1.280 0.20055
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) PhonLv IWFALS
PhonLev -0.907
IsWordFALSE -0.647 0.690
PhL:IWFALSE 0.640 -0.705 -0.983A frequentist regression model: plot predictions
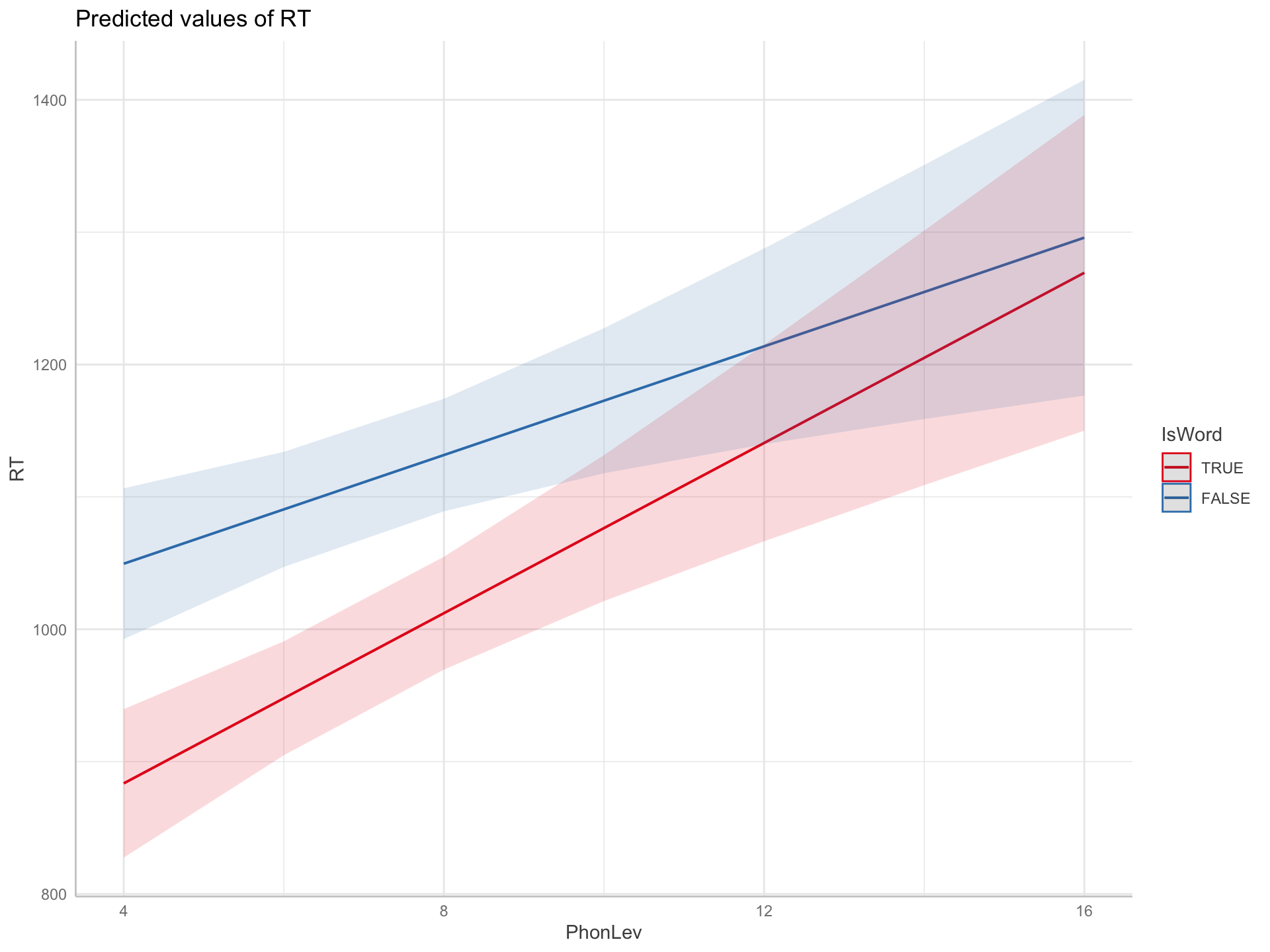
Figure 2: Predicted Reaction Times based on a frequentist regression model.
Increase model complexity
boundary (singular) fit: see help('isSingular')Warning: Model failed to converge with 1 negative eigenvalue: -2.5e+01
Let’s go Bayesian!

A Bayesian regression model
A Bayesian regression model: summary
Family: gaussian
Links: mu = identity; sigma = identity
Formula: RT ~ PhonLev + IsWord + PhonLev:IsWord + (PhonLev * IsWord | Subject)
Data: mald (Number of observations: 3000)
Draws: 4 chains, each with iter = 4000; warmup = 2000; thin = 1;
total post-warmup draws = 8000
Multilevel Hyperparameters:
~Subject (Number of levels: 30)
Estimate Est.Error l-95% CI u-95% CI Rhat
sd(Intercept) 89.60 32.39 23.91 163.00 1.00
sd(PhonLev) 6.81 4.93 0.32 17.93 1.00
sd(IsWordFALSE) 112.59 40.17 51.48 214.51 1.00
sd(PhonLev:IsWordFALSE) 6.49 5.27 0.22 19.83 1.00
cor(Intercept,PhonLev) -0.31 0.44 -0.90 0.67 1.00
cor(Intercept,IsWordFALSE) 0.43 0.29 -0.22 0.89 1.00
cor(PhonLev,IsWordFALSE) -0.19 0.44 -0.87 0.74 1.00
cor(Intercept,PhonLev:IsWordFALSE) -0.02 0.45 -0.82 0.82 1.00
cor(PhonLev,PhonLev:IsWordFALSE) -0.08 0.45 -0.84 0.78 1.00
cor(IsWordFALSE,PhonLev:IsWordFALSE) -0.30 0.47 -0.95 0.70 1.00
Bulk_ESS Tail_ESS
sd(Intercept) 928 371
sd(PhonLev) 750 1760
sd(IsWordFALSE) 1190 806
sd(PhonLev:IsWordFALSE) 1175 862
cor(Intercept,PhonLev) 2348 3006
cor(Intercept,IsWordFALSE) 1012 661
cor(PhonLev,IsWordFALSE) 811 1085
cor(Intercept,PhonLev:IsWordFALSE) 5085 3160
cor(PhonLev,PhonLev:IsWordFALSE) 3771 3295
cor(IsWordFALSE,PhonLev:IsWordFALSE) 1816 1348
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 755.02 48.28 659.98 848.85 1.00 3920 5309
PhonLev 31.99 6.52 18.99 44.79 1.00 4604 5222
IsWordFALSE 208.23 67.99 75.34 341.44 1.00 3642 4658
PhonLev:IsWordFALSE -11.06 9.12 -28.68 6.71 1.00 3743 4748
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 320.31 4.16 312.30 328.40 1.00 7998 5470
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).A Bayesian regression model: plot predictions
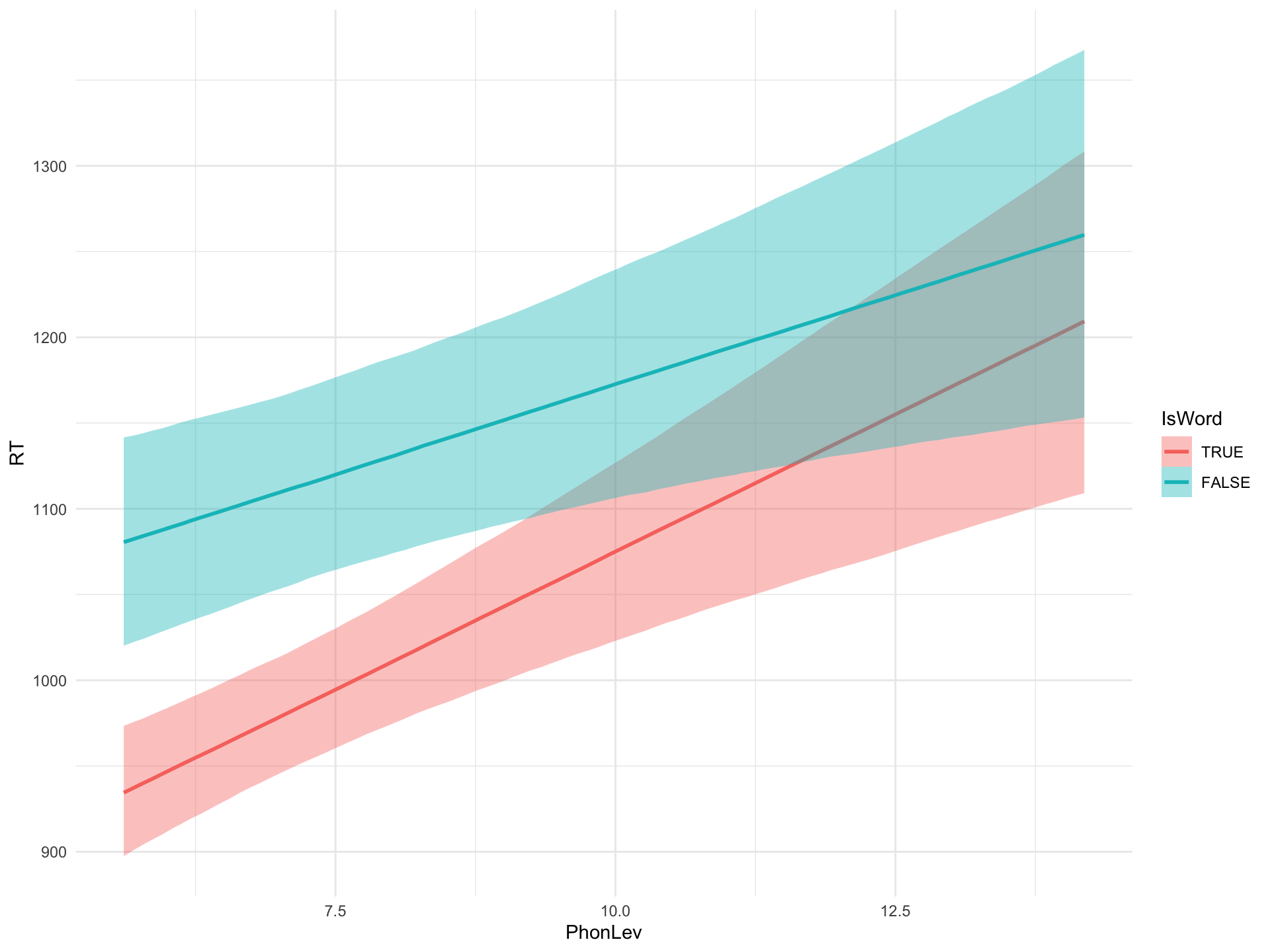
Figure 3: Predicted RTs based on a Bayesian regression model.
Let’s start small: A Gaussian model
\[ RT \sim Gaussian(\mu, \sigma) \]
Reaction Times \(RT\)
distributed according to \(\sim\)
a Gaussian distribution \(Gaussian()\)
with mean \(\mu\) and standard deviation \(\sigma\).
We start with a Gaussian model for pedagogical reasons. You would usually start with the model you need to answer your research questions.
Uncertainty of parameters
\[ \begin{align} RT & \sim Gaussian(\mu, \sigma)\\ \mu & \sim \mathcal{P}(\mu_1, \sigma_1)\\ \sigma & \sim \mathcal{P}(\mu_2, \sigma_2) \end{align} \]
Uncertainty can be expressed with probability distributions. We use \(\mathcal{P}\) as a generic (undetermined) probability distribution.
The Gaussian model estimates the probability distributions \(\mathcal{P}\) of \(\mu\) and \(\sigma\) from the data.
These probability distributions \(\mathcal{P}\) are called posterior probability distributions.
They can be summarised with \(\mu_1, \sigma_1, \mu_2, \sigma_2\).
Gaussian model of RTs
Try it yourself!
The model estimates the probability distributions of \(\mu\) and \(\sigma\).
To do so, a sampling algorithm is used (Markov Chain Monte Carlo, MCMC).
MCMC what?
Markov Chain Monte Carlo
Technicalities
You can find out how many cores your laptop has with parallel::detectCores().
The model summary
Family: gaussian
Links: mu = identity; sigma = identity
Formula: RT ~ 1
Data: mald (Number of observations: 3000)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 1046.57 6.38 1034.21 1058.95 1.00 3165 2707
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 347.57 4.52 338.77 356.50 1.00 3804 2833
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Interpret the summary: \(\mu\)
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 1046.57 6.38 1034.21 1058.95 1.00 3165 2707Intercept is \(\mu\).
\[ \begin{align} RT & \sim Gaussian(\mu, \sigma)\\ \mu & \sim \mathcal{P}(1046.57, 6.38)\\ \end{align} \]
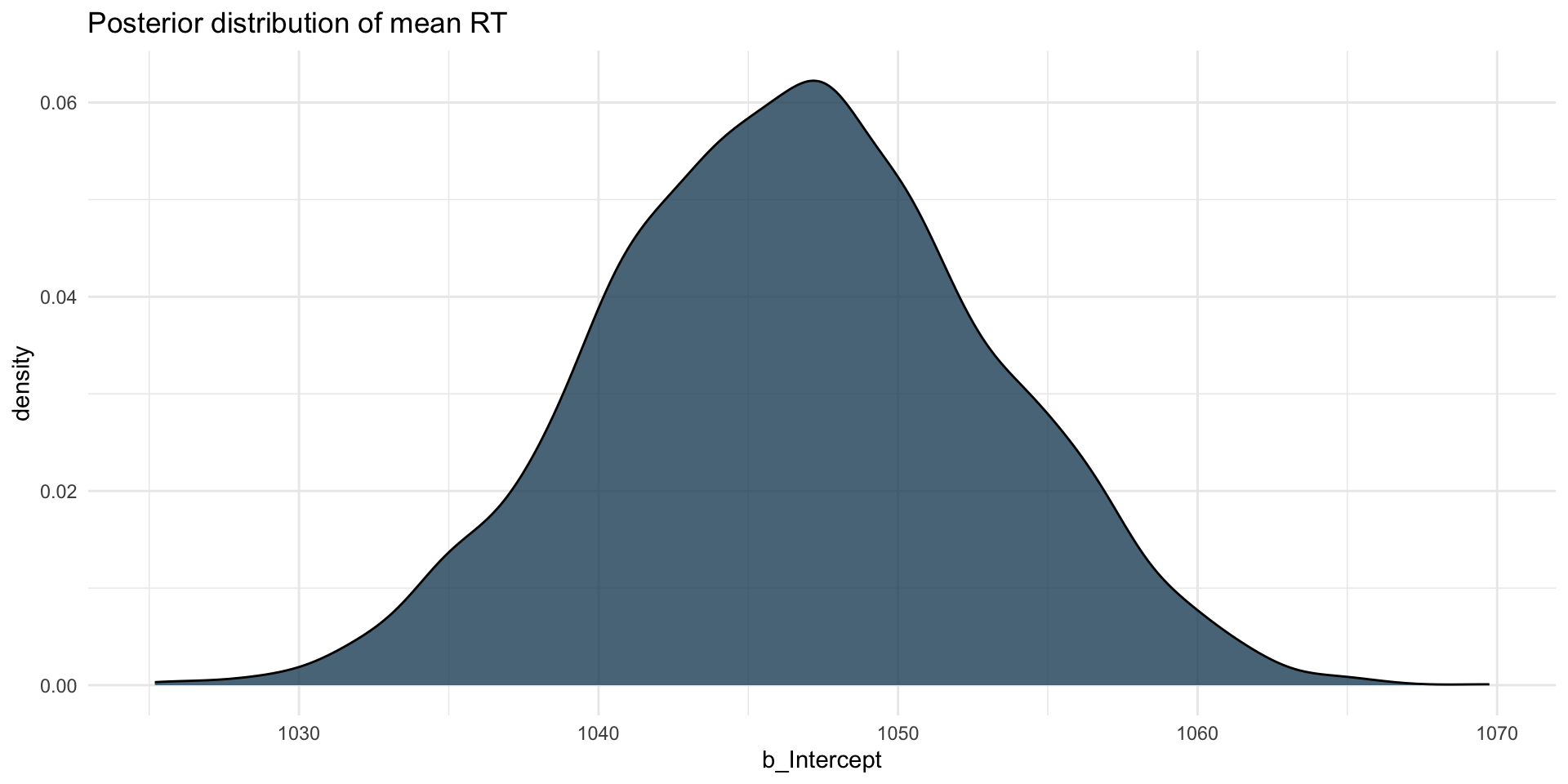
The posterior probability distribution of \(\mu\), i.e. of the mean RTs, is \(\mathcal{P}(1047, 6)\).
There is a 95% probability that the mean RTs is between 1034 and 1059 ms, conditional on the model and data. This is the 95% Credible Interval (CrI) of \(\mathcal{P}(1047, 6)\).
Interpret the summary: \(\sigma\)
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 347.57 4.52 338.77 356.50 1.00 3804 2833sigma is \(\sigma\).
\[ \begin{align} RT & \sim Gaussian(\mu, \sigma)\\ \mu & \sim \mathcal{P}(1046.57, 6.38)\\ \sigma & \sim \mathcal{P}(347.57, 4.52) \end{align} \]
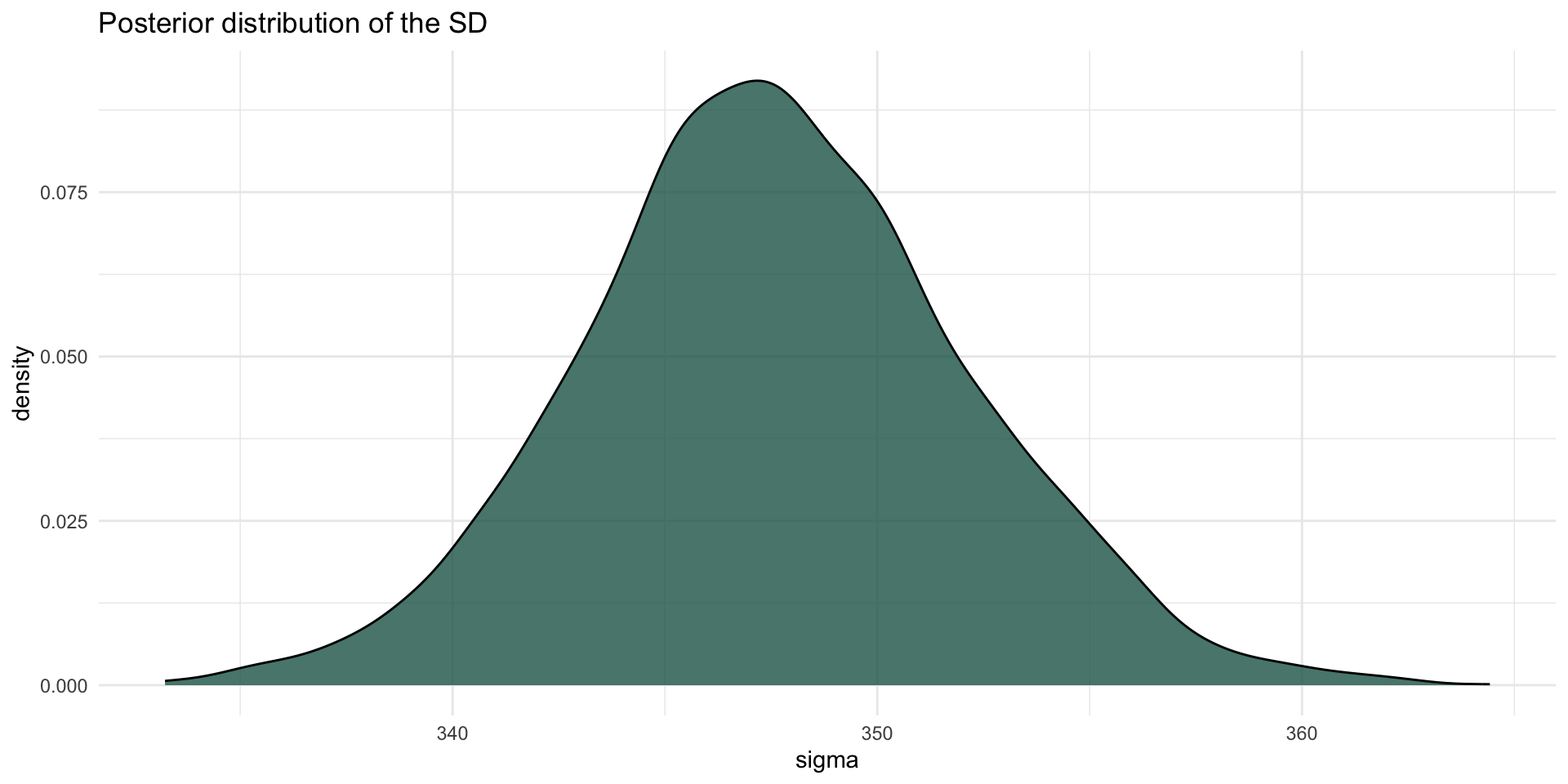
The posterior probability distribution of \(\sigma\), i.e. of the standard deviation of RTs, is \(\mathcal{P}(348, 5)\).
There is a 95% probability that the RT standard deviation is between 339 and 357 ms, conditional on the model and data.
Posterior probabilities
- These are posterior probability distributions.
- They are always conditional on model and data.
- The summary reports 95% Credible Intervals (CrIs), but there is nothing special about 95% and you should obtain a set of intervals.
Model plot
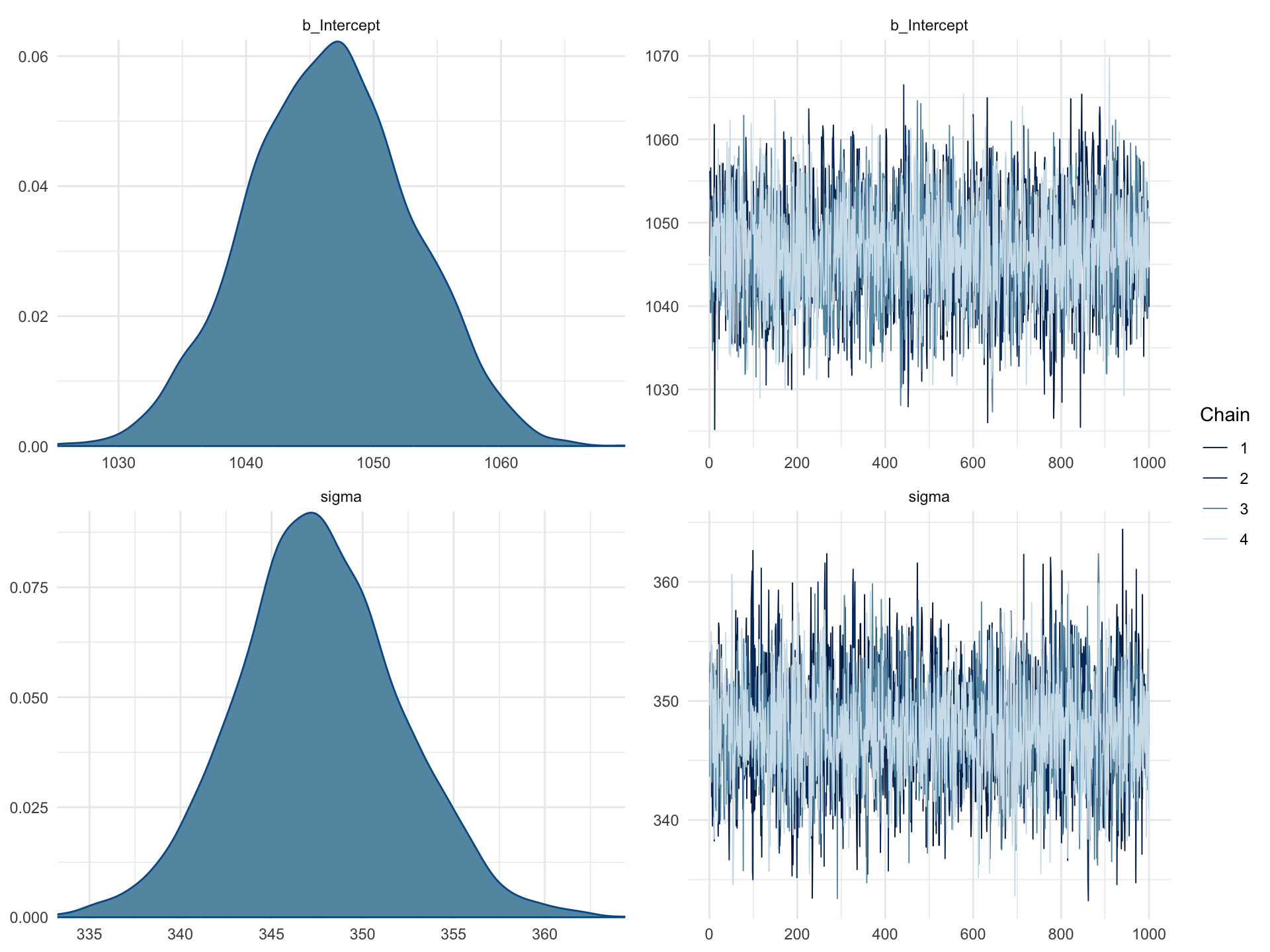
Figure 4: Density and trace plot of the Bayesian Gaussian model brm_2.
Extract the MCMC draws
# A draws_df: 1000 iterations, 4 chains, and 5 variables
b_Intercept sigma Intercept lprior lp__
1 1046 348 1046 -13 -21816
2 1052 353 1052 -13 -21817
3 1039 349 1039 -13 -21817
4 1045 343 1045 -13 -21817
5 1046 344 1046 -13 -21817
6 1050 348 1050 -13 -21816
7 1050 348 1050 -13 -21816
8 1044 345 1044 -13 -21816
9 1048 350 1048 -13 -21816
10 1036 345 1036 -13 -21818
# ... with 3990 more draws
# ... hidden reserved variables {'.chain', '.iteration', '.draw'}Get parameters
To list all the names of the parameters in a model, use:
Posterior distribution: mean RT
Now you can plot the probability density of the draws using ggplot2.
Posterior distribution: mean RT
Posterior distribution: sigma
Posterior distribution: sigma
Summary
Bayesian statistics is a framework for doing inference (i.e. learning something about a population from a sample).
The Bayesian framework is based on estimating uncertainty with probability distributions on parameters.
The syntax you know from
[g]lm[er]()works withbrm()from the brms package.Results allow you to make statements like “There is a 95% probability that the difference between A and B is between q and w”.