02 - Priors
Vowel duration
\[ \begin{align} vdur & \sim Gaussian(\mu, \sigma)\\ \end{align} \]
Pick a \(\mu\) and \(\sigma\).
Report them here: https://forms.gle/HRzH5CworngWrBv16.
Proposed distributions of vowel duration

Bayesian belief update
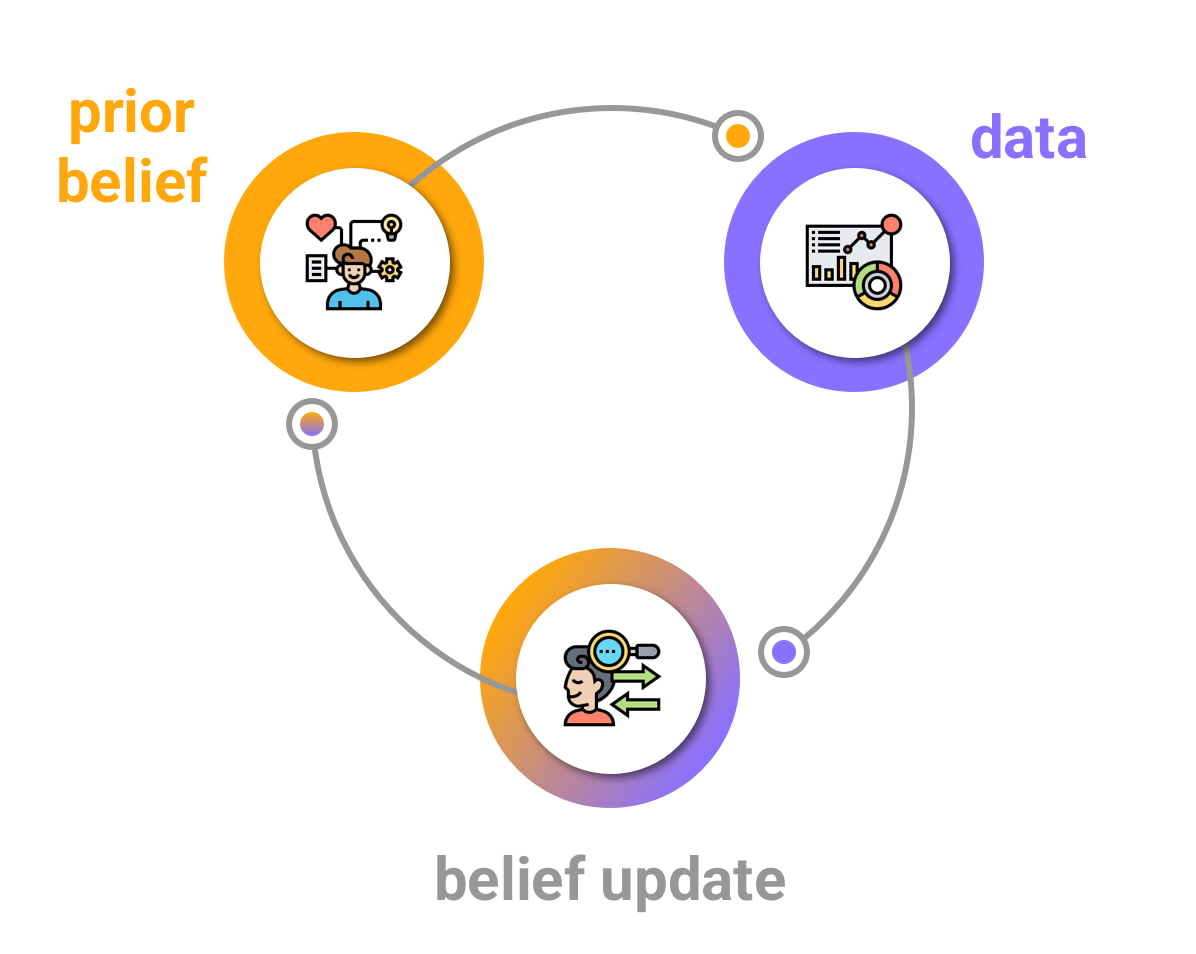
Priors
\[ \begin{align} vdur & \sim LogNormal(\mu, \sigma)\\ \mu & \sim Gaussian(\mu_1, \sigma_1)\\ \sigma & \sim Cauchy_{+}(0, \sigma_2)\\ \end{align} \]
Get default priors
Prior probability distributions

The empirical rule

The empirical rule

The empirical rule

The empirical rule

The empirical rule

Priors of the model
\[ \begin{align} vdur & \sim LogNormal(\mu, \sigma)\\ \mu & \sim Gaussian(\mu_1, \sigma_1)\\ \sigma & \sim Cauchy_{+}(0, \sigma_2)\\ \end{align} \]
Let’s pick \(\mu_1\) and \(\sigma_1\).
We can use the empirical rule.
Prior for \(\mu\)
\[ \begin{align} vdur & \sim LogNormal(\mu, \sigma)\\ \mu & \sim Gaussian(\mu_1, \sigma_1)\\ \sigma & \sim Cauchy_{+}(0, \sigma_2)\\ \end{align} \]
Let’s say that the mean vowel duration is between 50 and 150 ms at 95% confidence. In logs, that would be
log(50) = 3.9andlog(150) = 5.Get \(\mu_1\)
mean(c(3.9, 5)) = 4.45
Get \(\sigma_1\)
(5 - 3.9) / 4 = 0.275
Prior for \(\mu\)
\[ \begin{align} vdur & \sim LogNormal(\mu, \sigma)\\ \mu & \sim Gaussian(\mu_1 = 4.45, \sigma_1 = 0.275)\\ \sigma & \sim Cauchy_{+}(0, \sigma_2)\\ \end{align} \]
Prior for \(\mu\): plot

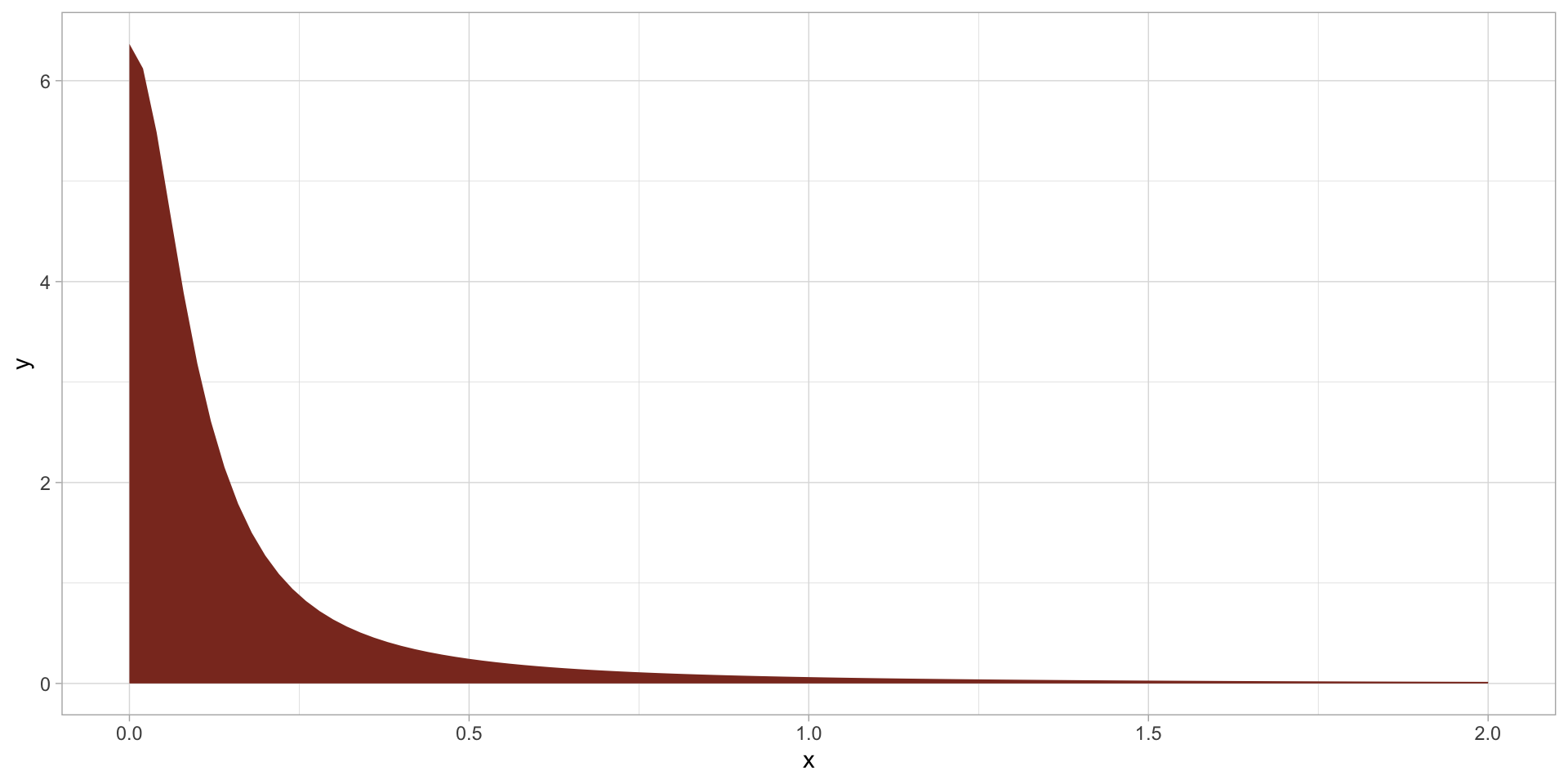
Prior for \(\sigma\)
Prior for \(\sigma\): plot
Prior predictive checks: sample prior
my_seed <- 3485
m_3_priors <- c(
prior(normal(4.45, 0.275), class = Intercept),
prior(cauchy(0, 0.1), class = sigma)
)
m_3_priorpp <- brm(
v1_duration ~ 1,
family = lognormal,
prior = m_3_priors,
data = token_measures,
sample_prior = "only",
cores = 4,
file = "data/cache/m_3_priorpp",
seed = my_seed
)Prior predictive checks: sample prior
Family: lognormal
Links: mu = identity; sigma = identity
Formula: v1_duration ~ 1
Data: token_measures (Number of observations: 1342)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 4.45 0.28 3.91 5.03 1.00 2736 1915
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.47 3.70 0.00 2.33 1.00 2902 2096
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Prior predictive checks: plot

Prior predictive checks: plot (zoom in)

Run the model
Model summary
Family: lognormal
Links: mu = identity; sigma = identity
Formula: v1_duration ~ 1
Data: token_measures (Number of observations: 1342)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-80% CI u-80% CI Rhat Bulk_ESS Tail_ESS
Intercept 4.59 0.01 4.58 4.61 1.00 4265 3162
Further Distributional Parameters:
Estimate Est.Error l-80% CI u-80% CI Rhat Bulk_ESS Tail_ESS
sigma 0.33 0.01 0.32 0.34 1.00 3407 2797
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Posterior predictive checks
