04 - Bernoulli models for binary outcomes
MALD: accuracy
Accuracy of responses: correctly or incorrectly recognised the word type (real or nonce-word).
Let’s model the effect of phonetic distance on accuracy.
Accuracy is a binary variable.
With binary variables we model the probability of one of the two levels. Here, correct.
The probability of one level of a binary variable follows a Bernoulli distribution.
Bernoulli model
\[ p(correct) \sim Bernoulli(p) \]
But… probabilities are bounded between 0 and 1.
For variables that can only have positive real numbers, we used the logarithmic function (log-normal models).
With probabilities, we can use the logistic function.
The logistic (or logit) function converts probabilities to log-odds (
qlogis()in R).
Logistic function
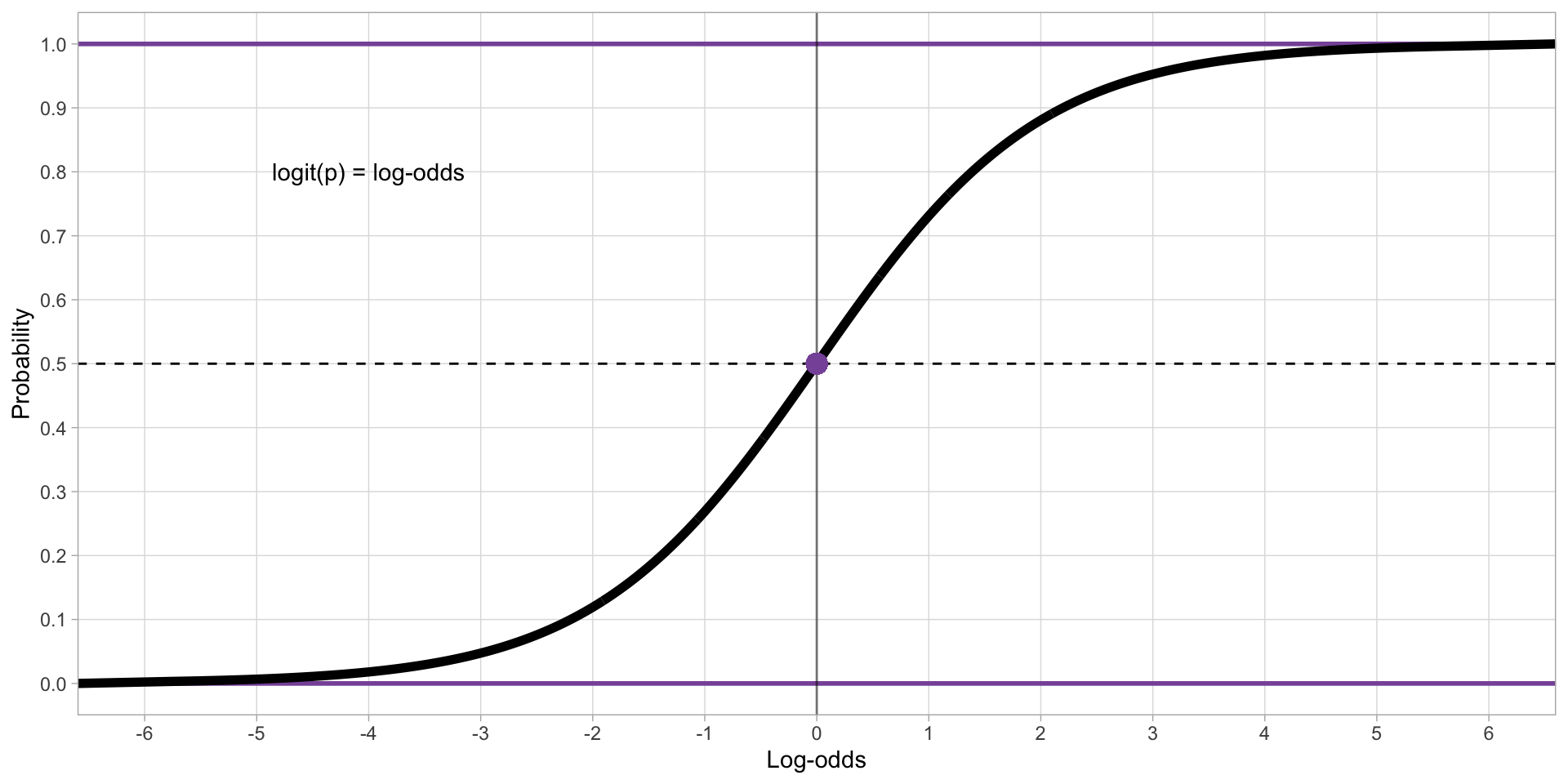
Figure 1: Correspondence between log-odds and probabilities.
Bernoulli (regression) models
Bernoulli models use the logistic function to treat probabilities as if they were unbounded.
The model’s estimates are in log-odds.
Log-odds can be converted back to probabilities with the inverse logit function (
plogis()in R).
- Bernoulli models are also known as binomial or logistic regression.
A Bernoulli regression of accuracy: code
A Bernoulli regression of accuracy: predicted accuracy
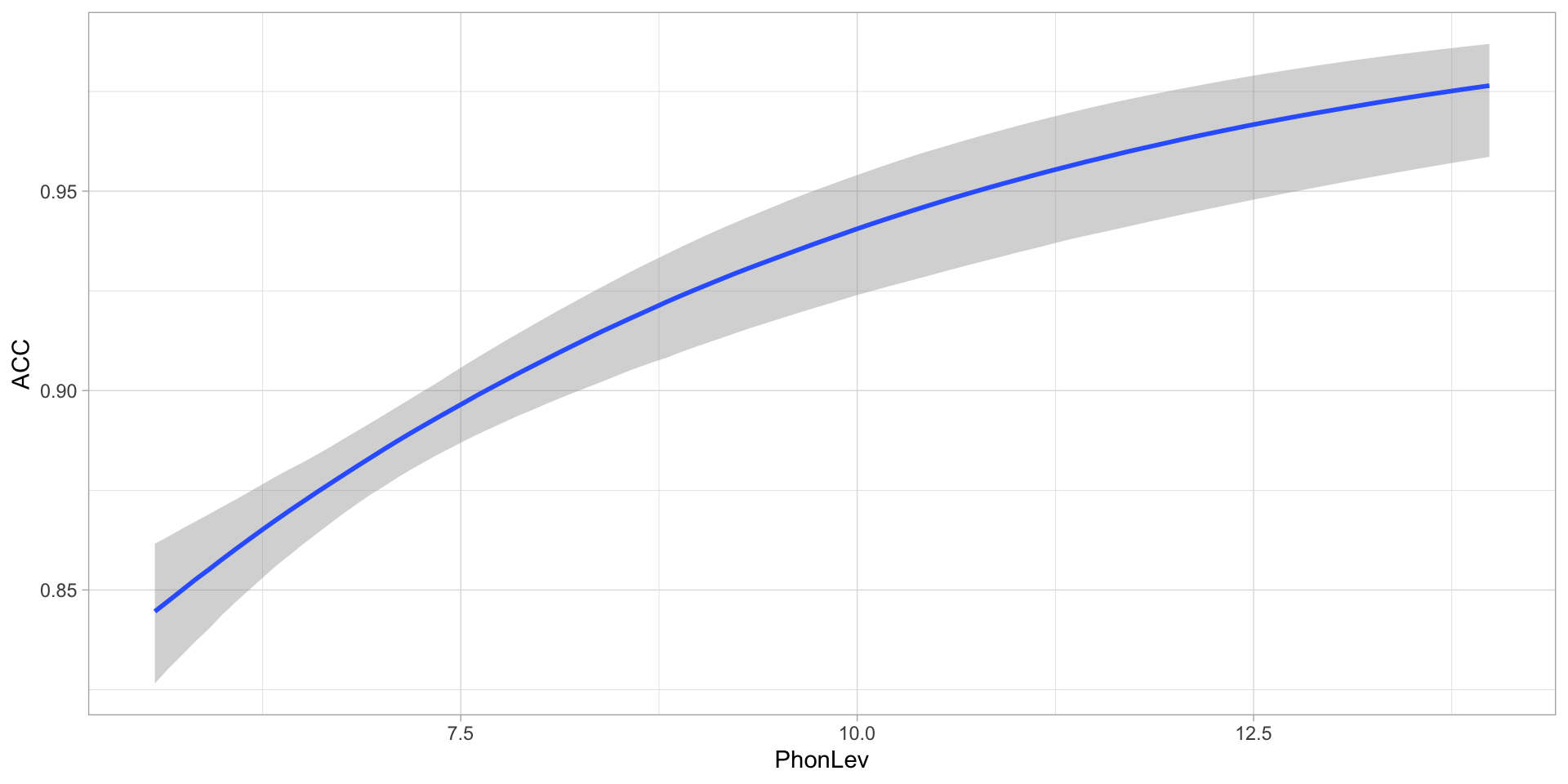
Figure 2: Predicted accuracy based on phonetic distance.
Brentari 2024: Lengua de Señas Nicaragüense (LSN)
# A tibble: 630 × 6
Group Participant Object Number Agency Num_Predicates
<chr> <dbl> <chr> <chr> <chr> <chr>
1 homesign 1 book single agent multiple
2 homesign 1 book single agent multiple
3 homesign 1 book plural no_agent multiple
4 homesign 1 coin single agent single
5 homesign 1 coin plural no_agent single
6 homesign 1 coin single no_agent single
7 homesign 1 coin single no_agent multiple
8 homesign 1 lollipop single agent single
9 homesign 1 lollipop single agent single
10 homesign 1 lollipop plural no_agent single
# ℹ 620 more rowsBrentari 2024
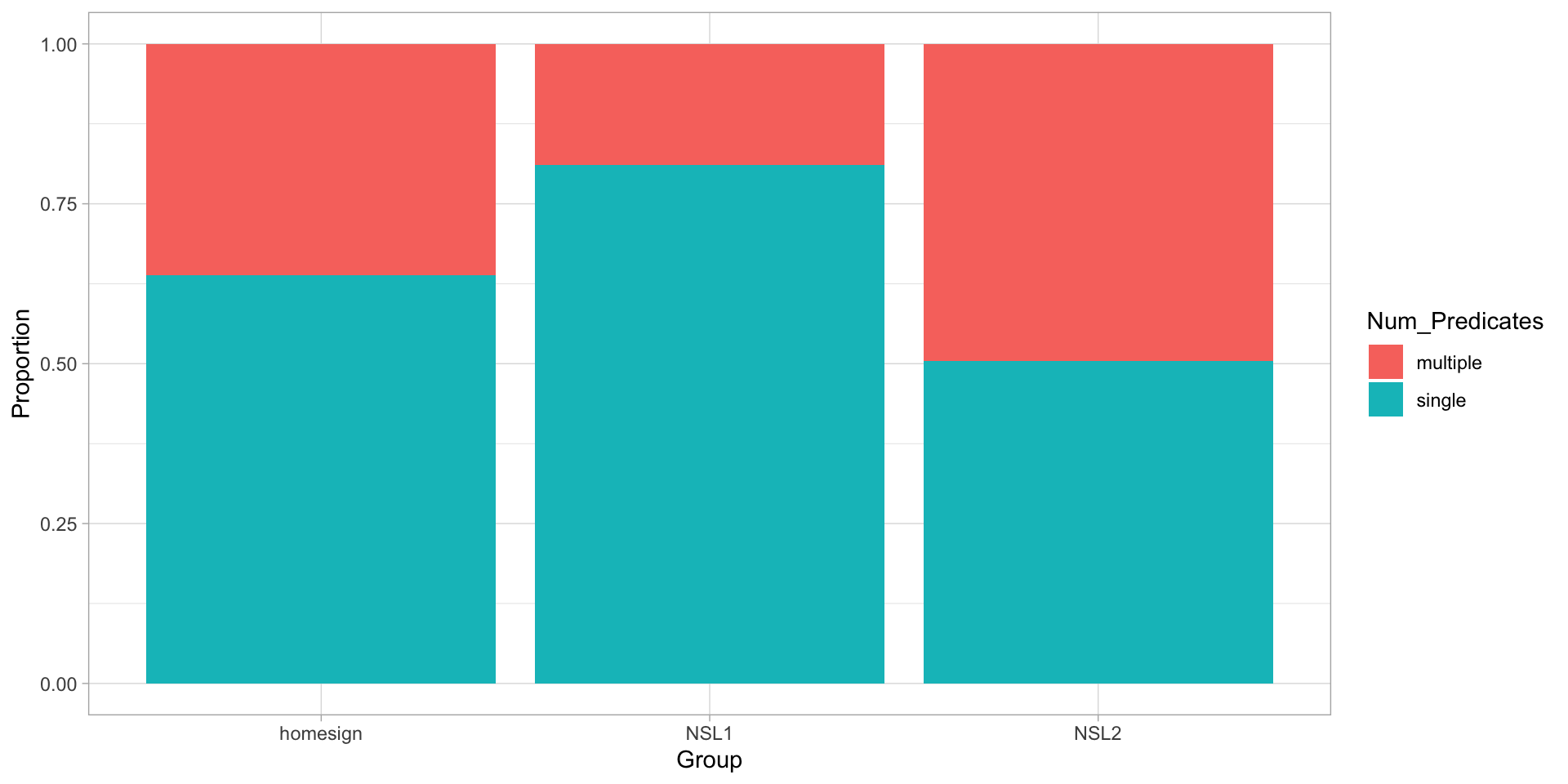
Figure 3: Proportion of predicate type by group.
A Bernoulli model of predicate type
\[ \begin{align} pred_i & \sim Bernoulli(p_i)\\ logit(p_i) & = \beta_1 \cdot G_{\text{HS}[i]} + \beta_2 \cdot G_{\text{NSL1}[i]} + \beta_3 \cdot G_{\text{NSL2}[i]} \end{align} \]
logit(p)indicates a logit function is used (so the estimates are in log-odds).\(G_{\text{HS}[i]}, G_{\text{NSL1}[i]},G_{\text{NSL2}[i]}\) are indicator variables, a way of including categorical predictors in a regression model.
Indicator variables
| \(G_{\text{HS}}\) | \(G_{\text{NSL1}}\) | \(G_{\text{NSL2}}\) | |
|---|---|---|---|
| Homesign | 1 | 0 | 0 |
| NSL1 | 0 | 1 | 0 |
| NSL2 | 0 | 0 | 1 |
\[ \begin{align} logit(p_i) & = \beta_1 \cdot G_{\text{HS}[i]} + \beta_2 \cdot G_{\text{NSL1}[i]} + \beta_3 \cdot G_{\text{NSL2}[i]}\\ \text{homesign, } logit(p_i) & = \beta_1 \cdot 1 + \beta_2 \cdot 0 + \beta_3 \cdot 0\\ & = \beta_1\\ \text{NSL1, } logit(p_i) & = \beta_1 \cdot 0 + \beta_2 \cdot 1 + \beta_3 \cdot 0\\ & = \beta_2\\ \text{NSL2, } logit(p_i) & = \beta_1 \cdot 0 + \beta_2 \cdot 0 + \beta_3 \cdot 1\\ & = \beta_3\\ \end{align} \]
Indicator variables
Note that indicator variables are dealt with by brms under the hood.
We talk about them to understand how the model is set up.
The model will estimate the probability \(p\) for each group.
But… the probability of what?
Prepare data
- The model will estimate the probability \(p\) of finding a multiple predicate, depending on group.
A Bernoulli model of predicate type: suppress the intercept
v1_duration ~ 1 + speech_rate_c
1stands for “intercept”. The model has an intercept and a slope.
Pred_Type ~ 0 + Group
0stands for “suppress the overall intercept”. The model estimates one “intercept” per level.
A Bernoulli model of predicate type: code
A Bernoulli model of predicate type: summary
Family: bernoulli
Links: mu = logit
Formula: Pred_Type ~ 0 + Group
Data: brentari2024 (Number of observations: 630)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Grouphomesign -0.57 0.16 -0.87 -0.26 1.00 4384 3351
GroupNSL1 -1.46 0.17 -1.79 -1.14 1.00 4209 2946
GroupNSL2 -0.02 0.14 -0.29 0.25 1.00 3865 2727
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).A Bernoulli model of predicate type: predicted probability
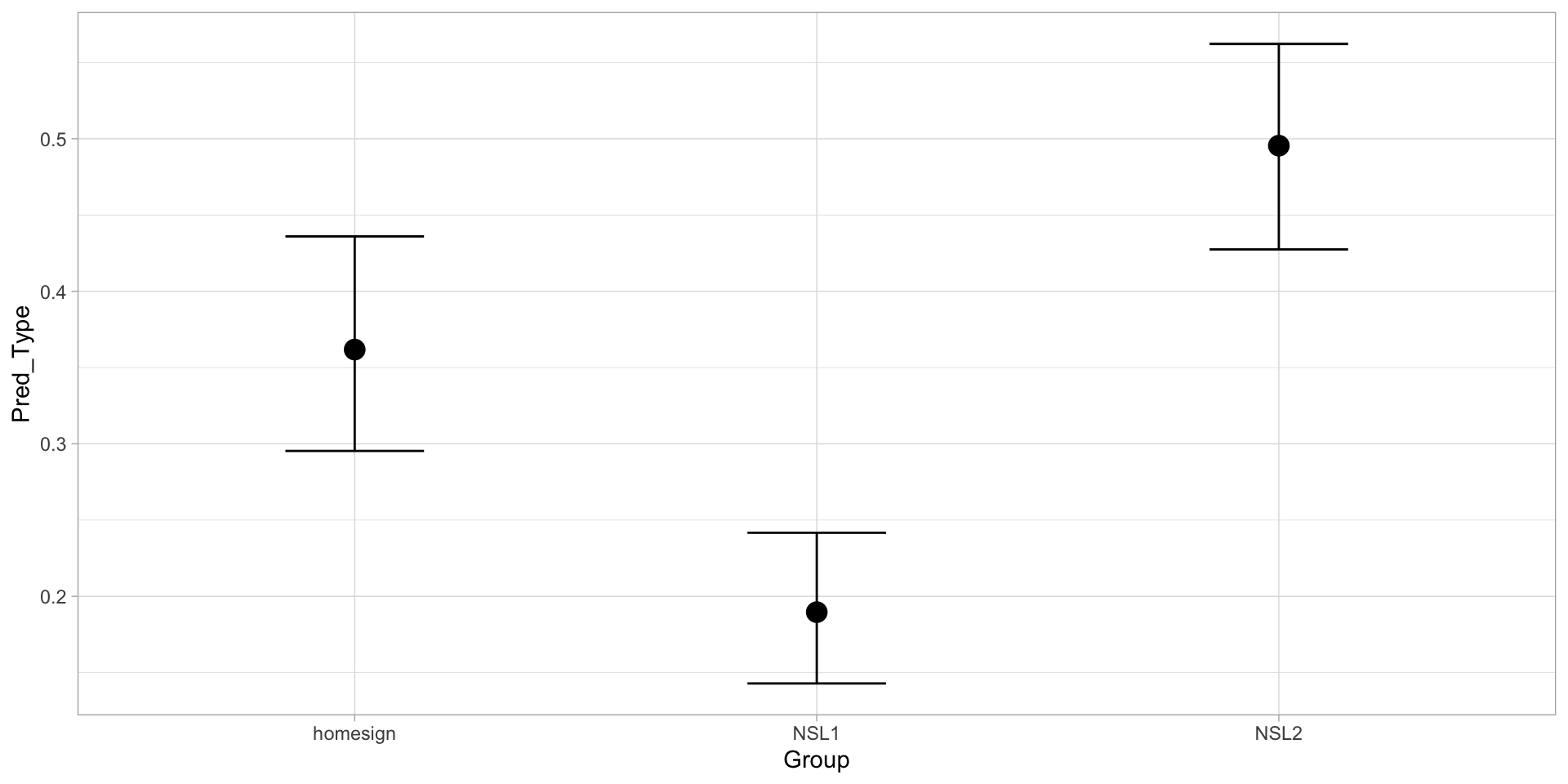
Figure 4: Predicted probability of multiple predicate by group.
Posterior draws
# A draws_df: 1000 iterations, 4 chains, and 5 variables
b_Grouphomesign b_GroupNSL1 b_GroupNSL2 lprior lp__
1 -0.43 -1.6 -0.112 0 -381
2 -0.17 -1.5 -0.049 0 -384
3 -0.81 -1.4 -0.205 0 -383
4 -0.52 -1.4 -0.110 0 -381
5 -0.74 -1.4 -0.175 0 -382
6 -0.21 -1.2 0.065 0 -384
7 -0.89 -1.3 -0.037 0 -383
8 -0.47 -1.3 0.095 0 -381
9 -0.56 -1.3 0.081 0 -381
10 -0.67 -1.5 0.048 0 -381
# ... with 3990 more draws
# ... hidden reserved variables {'.chain', '.iteration', '.draw'}Credible intervals: log-odds
library(posterior)
pred_bern_draws |>
pivot_longer(b_Grouphomesign:b_GroupNSL2, names_to = "coef", values_to = "est") |>
group_by(coef) |>
summarise(
ci_lo = quantile2(est, probs = 0.025),
ci_hi = quantile2(est, probs = 0.975)
)# A tibble: 3 × 3
coef ci_lo ci_hi
<chr> <dbl> <dbl>
1 b_GroupNSL1 -1.79 -1.14
2 b_GroupNSL2 -0.292 0.250
3 b_Grouphomesign -0.870 -0.257Credible intervals: probabilities
pred_bern_draws |>
pivot_longer(b_Grouphomesign:b_GroupNSL2, names_to = "coef", values_to = "est") |>
group_by(coef) |>
summarise(
ci_lo = round(quantile2(plogis(est), probs = 0.025), 2),
ci_hi = round(quantile2(plogis(est), probs = 0.975), 2)
)# A tibble: 3 × 3
coef ci_lo ci_hi
<chr> <dbl> <dbl>
1 b_GroupNSL1 0.14 0.24
2 b_GroupNSL2 0.43 0.56
3 b_Grouphomesign 0.3 0.44What about comparing the groups?
What if we want to know the difference between each group?
We can use the posterior draws. Simply take the row-wise difference and you will get a list of differences.
These are the posterior differences.
Comparing groups
pred_bern_draws <- pred_bern_draws |>
mutate(
NSL1_homesign = b_GroupNSL1 - b_Grouphomesign,
NSL2_homesign = b_GroupNSL2 - b_Grouphomesign,
NSL2_NSL1 = b_GroupNSL2 - b_GroupNSL1
)
pred_bern_draws# A draws_df: 1000 iterations, 4 chains, and 8 variables
b_Grouphomesign b_GroupNSL1 b_GroupNSL2 lprior lp__ NSL1_homesign
1 -0.43 -1.6 -0.112 0 -381 -1.13
2 -0.17 -1.5 -0.049 0 -384 -1.29
3 -0.81 -1.4 -0.205 0 -383 -0.64
4 -0.52 -1.4 -0.110 0 -381 -0.86
5 -0.74 -1.4 -0.175 0 -382 -0.71
6 -0.21 -1.2 0.065 0 -384 -1.04
7 -0.89 -1.3 -0.037 0 -383 -0.44
8 -0.47 -1.3 0.095 0 -381 -0.85
9 -0.56 -1.3 0.081 0 -381 -0.78
10 -0.67 -1.5 0.048 0 -381 -0.83
NSL2_homesign NSL2_NSL1
1 0.32 1.4
2 0.12 1.4
3 0.61 1.2
4 0.41 1.3
5 0.56 1.3
6 0.28 1.3
7 0.85 1.3
8 0.57 1.4
9 0.64 1.4
10 0.72 1.6
# ... with 3990 more draws
# ... hidden reserved variables {'.chain', '.iteration', '.draw'}Credible Intervals of the difference: log-odds
pred_bern_draws |>
pivot_longer(NSL1_homesign:NSL2_NSL1, names_to = "coef", values_to = "est") |>
group_by(coef) |>
summarise(
ci_lo = round(quantile2(est, probs = 0.025), 2),
ci_hi = round(quantile2(est, probs = 0.975), 2)
)# A tibble: 3 × 3
coef ci_lo ci_hi
<chr> <dbl> <dbl>
1 NSL1_homesign -1.34 -0.46
2 NSL2_NSL1 1.03 1.87
3 NSL2_homesign 0.11 0.95Credible Intervals of the difference: probabilities
pred_bern_draws |>
mutate(
NSL1_homesign_p = plogis(b_GroupNSL1) - plogis(b_Grouphomesign),
NSL2_homesign_p = plogis(b_GroupNSL2) - plogis(b_Grouphomesign),
NSL2_NSL1_p = plogis(b_GroupNSL2) - plogis(b_GroupNSL1)
) |>
pivot_longer(NSL1_homesign_p:NSL2_NSL1_p, names_to = "coef", values_to = "est_p") |>
group_by(coef) |>
summarise(
ci_lo = round(quantile2(est_p, probs = 0.025), 2),
ci_hi = round(quantile2(est_p, probs = 0.975), 2)
)# A tibble: 3 × 3
coef ci_lo ci_hi
<chr> <dbl> <dbl>
1 NSL1_homesign_p -0.26 -0.09
2 NSL2_NSL1_p 0.22 0.39
3 NSL2_homesign_p 0.03 0.23Summary
Binary variables can be modelled with a Bernoulli distribution.
Bernoulli models estimate the probability \(p\) of the second level in the variable.
Categorical predictors are modelled using indexing variables.
You can obtain posterior differences between levels by taking the difference of the posterior draws of those levels.